Expression Regulation of Starch and Storage Protein Synthesis Related Genes in Rice Grains
2019-02-18chengqingAN
, , , , e , , , , chen ,gqing , , , , , , , , , , , AN
1. College of Life Sciences and Institute for Conservation and Utilization of Agro-bioresources in Dabie Mountains, Xinyang Normal University, Xinyang 464000, China; 2. School of Science and Technology, Xinyang University, Xinyang 464000, China; 3. International Education College, Xinyang Normal University, Xinyang 464000, China; 4. Xinyang Academy of Agricultural Sciences, Xinyang 464000, China; 5. Xinyang Station of Plant Projection and Inspection, Xinyang 464000, China
Abstract Starch and the storage proteins are the main nutritious substances in crop grains, and their composition and content in grains play a decisive role in the grain quality of rice and other staple food crops. This review has mainly summarized the new advances in the expression regulation of starch and storage protein synthesis related genes in rice grains. Moreover, the challenges of the starch and storage protein synthesis substances in rice genetic improvement were also discussed. This review will provide important information for genetic improvement of grain quality in rice and, potentially, other staple cereals.
Key words Rice, Starch, Storage protein, Synthesis, Gene, Expression regulation
1 Introduction
Rice (OryzasativaL.) is one of the most important food crops, providing approximately 25% of energy demand to more than 3 billion people worldwide[1-3]. In the past few decades, in terms of grain improvement and cultivation of new rice varieties, the breeding programs of high-yield breeding, super-high-yield breeding, super-rice breeding and green super-rice breeding have been implemented, among which the increase of rice yield has always been an extremely important breeding target[4]. In the breeding process, the rice yield has been significantly improved. However, the genetic improvement of rice grain quality has not been improved simultaneously. As a result, the quality of rice grains on the market is not optimistic. However, the quality traits of rice grains are very complex quantitative traits, mainly composed of grain appearance and milling, eating, cooking, and nutritional qualities[5-7]. There is a certain degree of interaction between different rice quality traits and external environmental factors[6-9]. It was estimated that by 2050, the global population would even exceed 9 billion[10-11]. And with the gradual improvement of the quality of life, for a long time in the future, the demand for high-quality rice will continue to grow strongly.
The vast majority of nutrients in rice grains are starch and storage proteins. The sum of their weight accounts for more than 90% of the dry weight of rice grains[12-13]. The starch in rice includes amylase and amylopectin, of which the structural composition and relative contents have an important influence on many quality traits of rice grains, such as appearance quality, processing quality, cooking quality, nutritional quality and eating quality[14]. The storage protein content and amino acid content in rice grains and their relative balance are the most important factors determining the nutritional quality of rice grains[3, 15]. In addition, the storage protein content in rice also affects its food quality, processing quality and appearance quality[16]. According to different separation and extraction methods and solubility, the storage proteins in rice grains can be divided into glutelin, prolamin, globulin and albumin, which provide humans with about 15% of protein sources[12]. In short, the composition and relative content of various nutrients in rice grains have a certain degree of influence on the quality traits of rice grains, such as appearance quality, nutritional quality, cooking quality and processing quality[17-18]. Therefore, the composition and relative content of nutrients in rice are closely related to human nutrition and health.
In the past decade, significant progress has been made in the study of the genetic of major nutrients in rice grains[6, 19-21], and a large number of genes have been separated and cloned, which participate in the regulation of the synthesis, metabolism and degradation of major nutrients such as starch and storage proteins in rice[3, 19, 22-23]. The main nutrients (such as starch and storage proteins) in rice grains are closely related to the quality traits of rice. Therefore, to clarify the synthesis, regulatory networks and genetic basis of major nutrients such as starch and storage proteins in rice grains has important theoretical significance and application value for the genetic improvement of rice quality in the future. This paper focuses on recent advances in the expression and regulation of starch and storage protein synthesis related genes in rice grains, and proposes strategies for genetic improvement of major nutrients in rice grains, in order to provide reference for the genetic improvement of rice quality and the breeding of high-quality rice varieties.
2 Starch synthesis related genes and their regulation in rice grains
Starch in rice grains is one of the most important sources of dietary nutrients in humans, and it (including amylose and amylopectin) accounts for 80%-90% of the dry weight of rice grains[15]. The structure of starch and the relative content of the components in rice grains are closely related to the quality of rice. In particular, it has an important influence on the cooking and eating quality of rice (such as amylose content, gelatinization temperature and gel consistency)[9, 24]. The synthesis of starch in rice grains is very complicated. A range of genes, gene families, and transcription factors involved in starch synthesis and metabolism play an important role in the synthesis of rice starch[25-27]. Various abiotic stresses have a certain degree of influence on the synthesis and regulation of rice starch[28]. Recently, the results of a number of in-depth studies on transcription factors have shown that transcription factors are involved in the expression regulation of genes related for starch synthesis. This in turn affects the composition and structure of the starch.
The starch synthesis process in rice grains requires multiple steps of enzymatic reaction. A series of genes involved in starch synthesis and its metabolism are involved in this process. In addition, the genes involved in starch synthesis have more alleles, so that a large number of isozymes are involved in the synthesis and metabolism of grain starch (Fig.1)[29-30]. At present, a large number of studies have shown that the enzymes involved in starch synthesis include ADP-glucose pyrophosphorylase (AGPase), granule bound starch synthetase (GBSS), soluble starch synthase (SSS), starch branching enzyme (SBE), starch debranching enzyme (DBE), starch phosphorylase (PHO), amylose isomerase, glucose 6-phosphate translocator (GPT), pullulanase (PUL),etc[29, 31-33]. Among them, AGPase is a key enzyme in rich starch synthesis, which is the rate-limiting enzyme in the ADP-glucose pathway of active glucose donors[17, 34]. AGPase is encoded by six genes.OsAGPS-1 andOsAGPS-2 (a,b) are responsible for encoding the small subunit of AGPase, andOsAPL-1,OsAPL-2,OsAPL-3 andOsAPL-4 together encode the large subunit of AGPase[35-36]. The expression of these six genes is highly correlated with the activity of AGPase. When the activity of AGPase is high, it can promote the synthesis of starch in rice grains, thus the grain weight increases[37]. Otherwise, the total starch content in rice grains decreases and the endosperm dries up[32, 38]. Therefore, by regulating the expression of theAGPasegenes, it is possible to control the activity of AGPase. Finally, the improvement of starch quality in rice grains can be achieved.
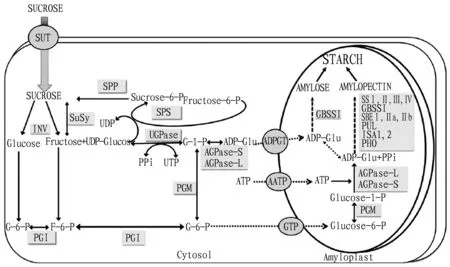
Note: SS, starch synthetase; GBSS, granule bound starch synthase; SBE, starch branching enzyme; PUL, pullulanase; ISA, isoamylase; PHO, starch phosphorylase; AGPase, ADP-glucose pyrophosphorylase; PGM, phosphoglucomutase; PGI, phosphoglucoisomerase; INV, invertase; SuSy, sucrose synthase; UGPase, UDP-glucose pyrophosphorylase; SPS, sucrose phosphate synthase; ADPGT, ADP-glucose transporter; AATP, plastidic ATP transporter; GTP, glucose-6-phosphate transporter.
Fig.1Simplifiedmodelofthestarchbiosyntheticpathwayinrice[30]
Another key enzyme responsible for starch synthesis in rice grains is starch synthetase (SS), which includes GBSS and SSS. Among them, GBSS-1 and GBSS-II are two isomerases of GBSS. GBSS-1 is encoded by theWaxygene (also known as theWxgene). In rice, it is mainly responsible for the synthesis of long-chain amylose in grains[39-40].Wxa,Wxb,Wxin,Wxg3,Wxmq,Wxmp,Wxop, andWxare all alleles of theWaxygene, which together directly or indirectly control the activity of starch synthase, and finally determine the amylose content in rice grains[17,25,41]. Soluble starch synthase has 8 kinds of isomerases in rice,SS-I,SS-II(a,b,c),SS-III(a,b) andSS-IV(a,b), among whichSS-Igene has not yet found the corresponding allelic form, andSS-Ienzyme plays an important role in the synthesis of short sugar chains synthesized by starch[42-43]. In the synthesis of rice starch, especially when the sugar chain of amylopectin is extended, the soluble starch synthase encoded bySS-II(a,b,c),SS-III(a,b) andSS-IV(a,b) genes is more active against ADP-glucose than GBSS. Therefore, they are beneficial to the synthesis and extension of amylopectin.SS-I,SS-IIaandSS-IIIagenes synergistically regulate the synthesis of amylopectin side chains in rice grains. TheSS-II(a,b,c) andSS-III(a,b) genes are all specifically expressed genes in rice. They are mainly concentrated in the endosperm or leaves of rice[17, 44]. Therefore, the genes encoding particle-coagulated starch synthase and soluble starch synthase and their corresponding isomerases play a key role in the synthesis and metabolism of amylose and amylopectin in rice grains.
In the synthesis and metabolism of starch in rice grains, starch branching enzyme (SBE) and starch debranching enzyme (DBE) are mainly responsible for the introduction of starch side chains and the removal of inappropriate side chains[45-46]. TheSBE-I,SBE-II(a,b) andSBE-IIIgenes respectively encode four isomerase corresponding to starch branching enzyme. They introduce α-1.6-glycosidic linkages in the backbone of starch glucan, resulting in the production and elongation of starch side chains[47-48]. Mutation or down-regulation ofSBE-1 gene has little effect on the grain appearance quality in rice. However, it can significantly increase the taste value of rice, and significantly reduce the gelatinization temperature of rice grains[18, 32]. TheSBE-IIagene is mainly responsible for the synthesis of short amylopectin, which further forms glucose chains on the amylopectin of different lengths. It may play an important role in the cultivation of new varieties of highly resistant starch in rice[47]. TheSBE-IIbgene is mainly responsible for the biosynthesis of the A chain in starch. It has high homology with theSBE-IIagene. However, there are obvious differences in the functions of the two, and the functions are not superimposed[30]. Starch debranching enzymes include isoamylase (ISA) and pullulanase (PUL). Among them, isoamylase is encoded by three genes,ISA1,ISA2 andISA3. TheISA1 andISA2 genes play an extremely important role in the synthesis process of amylopectin in rice endosperm. If these two genes are mutated, it will lead to the inability to synthesize starch in rice endosperm[33]. TheISA3 gene has little effect on the stored starch in rice grains. Instead, it is mainly responsible for the metabolic process of transient starch in seeds[49]. Mutation or loss of function of thePULgene will result in a significant increase in short-chain branched starch in rice endosperm[50-51]. Therefore, in the genetic improvement process of starch in rice grains and the corresponding rice quality, specific types of alleles need to be screened based on different breeding objectives, thereby speeding up the breeding of new rice varieties.
At present, the functional studies of individual genes related to starch anabolism in rice grains are relatively thorough. Multiple starch synthesis related genes jointly control the metabolic process of starch in grains[13]. In addition, a large number of starch synthesis related transcription factors are involved in the synthesis of grain starch. Mutation of one of these genes will cause changes in other genes involved in starch metabolism and their transcription[52]. Thus, a highly complex regulatory network of starch synthesis related genes and their transcription factors is formed in rice grains. However, little is known about how to coordinate and regulate expression between them. For example, the expression product ofDu1 gene can regulate the splicing mode and efficiency ofWxgenes. Finally, it directly affects the content of amylose in rice grains[53-54]. RSR1 (Rice Starch Regulator 1) is a transcription factor of the APETALA2/ethylene-response element binding protein family[55]. If the expression level ofRSR1 gene is increased, the starch synthesis related genes in rice grains are inhibited. Another transcription factor,OsbZIP58, binds directly to promoters of multiple starch anabolism-related genes (such asSSSIIa,Wx,AGPL3,SBEI,SBEIIbandISA2etc.), or interacts withOsLOL1[23, 56]to regulate the expression of the above genes, ultimately affecting the accumulation of starch, the formation of chalkiness and the germination of rice seeds. In addition, some other transcription factors are also involved in the synthesis of amylose or amylopectin in rice grains, such asOsbZIP33,OsBP-5 andFLOURYENDOSPERM2[56-58]. They provide new strategies for the genetic improvement of the starch content and its composition in rice grains. Therefore, in the future, the research on the synergistic regulation of starch synthesis related genes and transcription factors in rice grains needs to be strengthened to gradually analyze the complex regulation network of synthesis and degradation of starch, thereby accelerating the improvement of rice starch.
3 Protein synthesis related genes and their regulation in rice grains
The content of storage proteins and starch in rice grain accounts for more than 90% of its dry weight. The content of proteins in rice grains does not exceed 16% of its dry weight[3, 59], but rice supplies about 15% of the world’s edible protein[12], which is the main source of protein for humans[3, 60]. The storage proteins in rice grains include glutelin, prolamin, globulin and albumin[12]. Among them, the most important ones are glutelin and prolamin, and glutelin is easily absorbed by human body. Therefore, the protein content in rice grains is an important factor in determining the nutritional quality of rice grains.
At present, a large number of research results show that the synthesis of proteins in rice grains requires the participation of multiple types of storage protein encoding related genes. These genes or gene families constitute a complex regulatory network[61-62]. Glutelin in rice grains is encoded by at least four glutelin genes,GluA,GluB,GluCandGluD. TheGluAhas at least 4 subtypes,GluA-1,GluA-2,GluA-3 andGluA-4. TheGluBhas at least 6 subtypes,GluB-1,GluB-2,GluB-3,GluB-4,GluB-5 andGluB-6. TheGluChas at least 2 subtypes. At present, only one subtype ofGluDhas been identified. All the types ofGluconstitute the glutelin gene family[63-64]. The prolamin in rice grains mainly contains three types, 16 kDa prolamin, 13 kDa prolamin and 10 kDa prolamin. The content of 13 kDa prolamin is relatively high, and it is further divided into three sub-categories, sub-category I, sub-category II and sub-category III[65]. This shows that similar to glutelin, the prolamin in rice grains is also encoded by multiple gene families. Each haplotype of the prolamin has more repetitions. As a result, studies on the prolamin-related genes in rice grains have lagged behind. The albumin and globulin in rice grains are mainly found in the aleurone layer of the embryo and the endosperm, and their contents are relatively low. At present, the genetic resource related to albumin and globulin cloning is still limited. Both theRA16 andRA17 genes encode albumin, belonging to the same albumin gene family. The similarity between the two gene sequences is also relatively high[66]. Recently, another gene,RAG2, encoding the albumin in rice grains, was isolated and cloned. It was found thatRAG2 is a gene specifically expressed in rice grains during filling stage; overexpression ofRAG2 can significantly increase protein content and grain size in rice grains, further increasing the yield and improving the grain quality in rice[19]. Reports on the isolation and cloning of rice globulin-related genes are rare, and only theGlbgene was successfully isolated and cloned[67]. Among the genes related to the storage proteins that have been cloned, most of them are found in rice mutants, such as the 11 mutants associated with rice glutelin synthesis:OsRab5a,gpa3Osvpe1,esp2,glup1 (esp5),glup2 (esp6),glup3 (esp7),glup4 (esp8),glup5,glup6 andglup7[62, 68]. Therefore, among the genes encoding storage proteins in rice grains, the most isolated and cloned genes are the ones belonging to the glutelin and prolamin gene families, and the genes encoding globulin and albumin are still relatively rare. If individual gene that encodes glutelin is mutated, there is often no significant effect on the amount of glutelin in the final rice grains, suggesting that the genes encoding the storage proteins in rice grains may have functional redundancy.
The synthesis of proteins in rice grains involves the regulation of the expression of a large number of gene families. Manycis-acting elements andtrans-acting elements are also involved in this process. Some commoncis-acting elements have been found in rice storage protein synthesis related genes and their gene family members. In theGluA,GluBandGluDgenes that constitute the glutelin gene family, some very conservedcis-acting elements such as ACGT box, prolamin box, GCN4 and AACA box can be found[69-70]. There are some relatively conserved elements in the genes encoding rice prolamin, such as the sequences similar to the GCN4-Box and prolamin box[8]. This implies that similar regulatory mechanisms may exist among the genes encoding rice storage proteins and their gene families in the process of synthesizing seed storage proteins.Trans-acting elements, especially transcription factors, are also involved in the process of protein synthesis and regulation in rice grains. For example, two transcription factors, OsRISBZ1 and OsRPBF, can regulate the expression of glutelin-related genes in rice grains, which in turn has an important effect on the content of storage proteins in rice grains[69,71]. RITA-1 protein and other members of the basic leucine zipper protein family (e.g. RISBZ1, RISBZ2, RISBZ3, RISBZ4 and RISBZ5), astrans-acting elements, can bind to GCN4-Box on the promoter of the genes involved in storage protein synthesis and synergistically regulate glutelin synthesis and its content in rice grains[69]. Thetrans-acting element RPBF not only recognizes the conservedcis-acting elements of theGluB-1 gene promoter. Moreover, it can synergistically regulate the expression of various storage protein genes together with RISBZ1 (e.g.GluB-1, 10-kDa prolamin gene, 13-kDa prolamin gene,GluD-1,GluA-1,GluA-2,GluA-3, 16-kDa prolamin gene and α-globulin gene)[69,71]. Therefore, protein synthesis in rice grains is controlled by members of multiple gene families, and manycis-acting elements andtrans-acting elements are involved in storage proteins synthesis. How these genes and their regulatory elements synergistically regulate the synthesis of storage proteins in rice grains till needs study in depth.
The protein content of rice grains is a quantitative trait[3]. Different research groups have conducted extensive research on the grain protein content in rice and made important progress. At present, different numbers of QTLs are detected on 12 chromosomes of rice genome to control the storage protein content in rice grains[64, 72-75], and some QTLs can be stably detected in different research groups and different ecological environments. For example, the region of rice chromosome 6 where theWaxygene is located has been found by many different research groups to be closely related to grain protein content in rice[73, 75-77]. Between RM472-RM104 on the long arm of rice chromosome 1, a QTL site with a large genetic effect can be detected to control the grain protein content in Wuhan and Hainan[78]. The genetic effect of the QTL was verified by constructing a near-isogenic line (NIL), and the QTL geneOsAAP6 was cloned by means of map-based cloning[3]. The further functional study showed that increasing the expression ofOsAAP6 gene in rice can promote the expression of genes related to storage protein synthesis in rice grains, leading to an increase in the levels of glutelin, prolamin, albumin and globulin in mature rice grains (Fig.2). At the same time, the synthesis of amylopectin will be severely inhibited. Therefore, the location of QTLs that control the protein content in rice grains and the isolation and cloning of QTL genes will lay a solid foundation for further revealing the mechanism and regulation network of natural variation of grain protein content in rice.
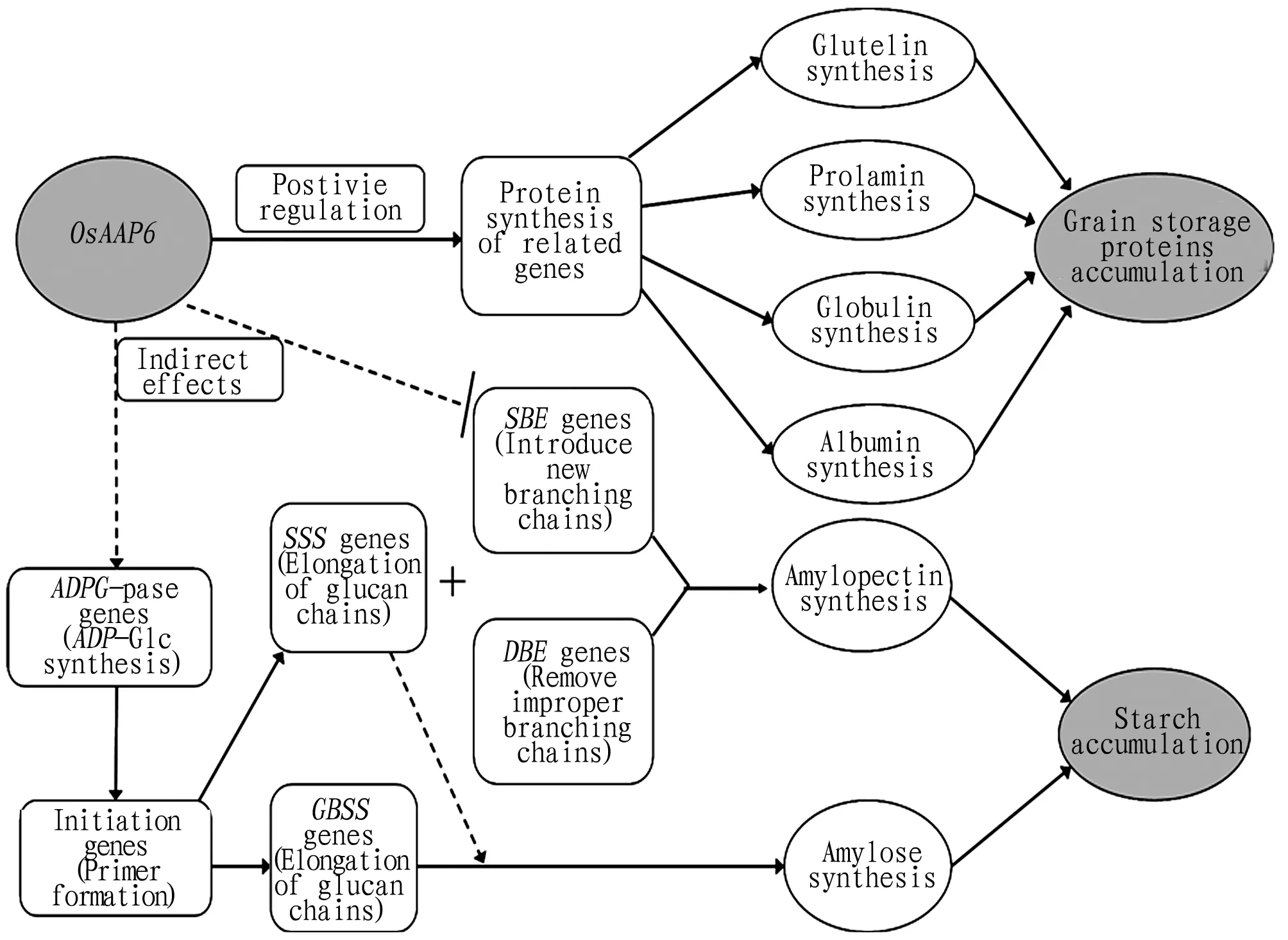
Note: ADPG-pase, ADP-glucose pyrophosphorylase; GBSS, granule-bound starch synthase; SSS, soluble starch synthase; SBE, starch branching enzyme; DBE, debranching enzyme.
Fig.2ThepossibleroleofOsAAP6geneingrainproteinsynthesis[3]
4 Strategies for improvement of main nutrients in rice grains
The main nutrients of rice grains include starch, storage proteins, lipids and amino acids. If these major nutrients want to be improved through genetic engineering or metabolic engineering, the genes of interest need to be driven by suitable promoter to produce the optimal functional proteins or enzymes[79], thereby promoting the synthesis of large and micronutrients in rice grains, instead of affecting other metabolic pathways and normal growth and development of plants[80]. Currently, the mechanism of biosynthesis of some nutrients in rice and its regulation is still unclear. As a result, the application of genetic engineering or metabolic engineering in rice genetic improvement is limited. Using multi-omics (proteomics, metabolomics, transcriptomics,etc.), a great deal of research has been done on nutrients in rice grains and important progress has been made. For example, some poor allergenic proteins in rice modified by genetic engineering can be screened by proteomics[81]; and metabolomics can accurately quantify multiple metabolites in rice, and identify metabolite markers associated with abiotic stress resistance and nutritional starvation in rice[82-83], which is particularly helpful in promoting the accumulation of beneficial nutrients in seeds. This will be of great significance for improving human nutrition and health.
In recent years, the rapid development of technologies such as gas chromatography-mass spectrometry, liquid chromatography-mass spectrometry, capillary electrophoresis-mass spectrometry, X-ray fluorescence spectroscopy, X-ray spectroscopy, scanning electron microscopy and transmission electron microscopy, as well as single-cell live imaging has provided new technical solutions for the in-depth study of the synthesis and accumulation of major nutrients in rice grains. However, for multiple steps in which multiple genes simultaneously act on the biosynthetic pathway, or for multiple traits of multiple synthetic pathways, simultaneous genetic improvement in rice grains remains challenging. Although the functions of many genes related to the synthesis of nutrients in rice grains have been revealed, functional genes are often directly or indirectly regulated by other genes. In addition, the phenomenon of one factor leading to many results is more common in genes. Therefore, functional genes that can be widely applied in rice grain nutrient genetic breeding are relatively rare. Moreover, development-specific, tissue-specific or expression-inducible promoters are often required to drive expression of these functional genes so as to facilitate the synthesis and accumulation of nutrients in rice grains.
For a long time, whether people can accept nutrient-fortified rice and its food has always been a hot issue in global discussions. Although no GM foods have been observed to adversely affect human health and the ecological environment, the commercial production and application of nutrient-fortified rice and its food will take time. In the past ten years, sequence-specific nuclease-based genome editing technology is evolving rapidly, and it has become one of the most effective new tools for grain genetic improvement in rice[84-85]. In particular, the CRISPR/Cas9 technology has simple operation, low cost, high induction efficiency, and the ability to obtain stable genetically modified plants. In addition, it is possible to simultaneously modify and edit genes at different positions within the rice genome[85-86]. Therefore, CRISPR/Cas9 technology has been widely used in the research of rice nutrient anabolism[87]. The genome editing technology represented by CRISPR/Cas9 technology will play an increasingly important role in the genetic improvement of rice grain nutrients and the cultivation of new varieties, and will greatly accelerate the genetic improvement of grain quality in rice and, potentially, other major grain crops.
杂志排行
Asian Agricultural Research的其它文章
- Possibility of Socio-economic Attributes and ICT Application in Improving Quality of Farmers
- Analysis of Gene Expression Profiles of Rice Mutant SLR1 Based on Microarray Data
- Optimization of Fermentation Process of Carrot Juice by Probiotics and Analysis of Aroma Components
- Safety Assessment and Distribution Characteristics of Heavy Metal Pollutants in Livestock and Poultry Meat from Different Regions of China
- Cucumber Downy Mildew: Research Progress and Registered Fungicides
- Effects of Different Seedling Raising Substrates on Quality of Rice Seedlings
