基于非培养手段的多环芳烃降解微生物解析
2018-09-25陈亚婷张娜娜连路宁
陈亚婷,姜 博,邢 奕,张娜娜,连路宁,路 培
基于非培养手段的多环芳烃降解微生物解析
陈亚婷,姜 博*,邢 奕*,张娜娜,连路宁,路 培
(北京科技大学能源与环境工程学院,工业典型污染物资源化处理北京市重点实验室,北京 100083)
本文对原位识别土壤未培养微生物的非培养手段进行综述,揭示其在PAHs降解中发挥的作用.首先对土壤中PAHs的降解微生物、降解原理以及功能基因进行全面总结;其次对荧光原位杂交技术(FISH)、稳定同位素探针技术(SIP)、宏基因组学技术(Metagenomics)、转录组学技术(Transcriptomics)、单细胞技术(Single-cell technology)以及磁性纳米材料技术(MMI)等非培养手段的原理、特点以及应用进行归纳总结.重点讨论这6种非培养手段在原位识别PAHs降解微生物中的应用潜力.其中SIP在研究PAHs降解机制中最为成熟、稳定,应用最广泛.MMI作为一种新型的非培养手段,能够从复杂的生态环境中分离获得活的功能微生物,为深入探究未培养的PAHs降解微生物提供解决途径.
未培养微生物;多环芳烃降解;土壤;功能微生物;非培养方法
土壤的多孔结构以及富含有机质、沉积物等特点,为PAHs的全球范围迁移提供了储存场所.这使得土壤中的PAHs含量高、难处理.寻找高效的土壤PAHs修复技术迫在眉睫.修复PAHs污染土壤的常用技术包括物理修复、化学修复以及生物修复[11].物理修复技术是根据污染物颗粒粒径、密度、分布大小及表面性质,借助物理手段将污染物从土壤中分离或去除,包括热修复法、换土法及电动修复法等[12-13].物理法能耗高,受土壤性质影响大,易破坏土壤结构与性质,不能彻底清除污染物质且容易造成二次污染.化学修复技术是利用土壤中污染物性质,向其中加入相应化学物质并经过一系列化学反应,最终实现污染物改变、分离或固化,包括土壤淋洗法、电化学修复法、化学氧化法、化学还原法等[14-15].修复过程中污染物迁移扩散、废气废水处理、成本消耗以及二次污染等是制约化学修复技术发展的主要因素.相比之下,生物修复技术具有低成本、高效率、小污染、操作简便等优势,是土壤修复领域研究重点之一[16].
生物修复技术包括微生物修复技术和植物修复技术,本文针对PAHs污染土壤微生物修复技术的作用原理、特点以及研究进展进行论述,对微生物修复技术中常用的分子生物学技术的优缺点、发展现状、适用范围进行讨论,旨在为从事污染场地修复的研究人员,提供微生物修复技术的理论指导.
1 多环芳烃污染场地的微生物降解
微生物修复技术[17]是微生物利用PAHs作为生长繁殖的碳源或能源,合成自身生命物质的同时以共代谢以及矿化作用的方式降解土壤中的PAHs.参与PAHs降解的微生物包括细菌、真菌和藻类,其中对细菌的研究和报道较多,包括假单胞菌属()[18]、红球菌属()[19]、气单胞菌属()[20]、分枝杆菌属()[21]、微球菌属()[22]、诺卡氏菌属()[22]、黄杆菌属()[23]、伯克氏菌属()[24]、鞘氨醇单胞菌属()[25]、芽孢杆菌属()[28-29]等(表1).
PAHs的降解首先是羟化双加氧酶对苯环邻位C原子的羟基化,随后在多种酶的作用下逐步催化降解.目前,对PAHs代谢途径的推测主要依赖于识别和分析PAHs代谢产物.不同种类的PAHs代谢途径不同,不同微生物对同种PAHs的代谢途径也不相同,而同种微生物在代谢PAHs的过程中也会存在多种途径.例如含有3个苯环的菲在sp. P1-1菌株降解过程中不仅发生3,4位C被羟基化,也会在1,2或者9,10位发生C被羟基化,在不同酶的作用下生成1,2-二羟菲或联苯甲酸等[26].PAHs的代谢过程中涉及微生物多种酶的合成及催化,这些酶的合成及功能发挥受到多种功能基因的表达调控.如表2所示,已发现来源不同菌株的PAHs降解功能基因包括、、、及等基因,它们编码的酶在PAHs代谢过程中发挥着不同的作用.如基因簇编码的酶可以将菲转化为顺-3,4-二羟基-3,4-二氢菲(cis-3,4-dihydroxy-3, 4- dihydrophenanthrene),基因簇编码的酶将1-羟基-2-萘酸转化为邻苯二甲酸酯[27];菌株NAH7质粒携带的基因簇编码的酶将萘转化为水杨酸,基因簇编码的酶将水杨酸转化为乙酰辅酶A和丙酮酸[28].
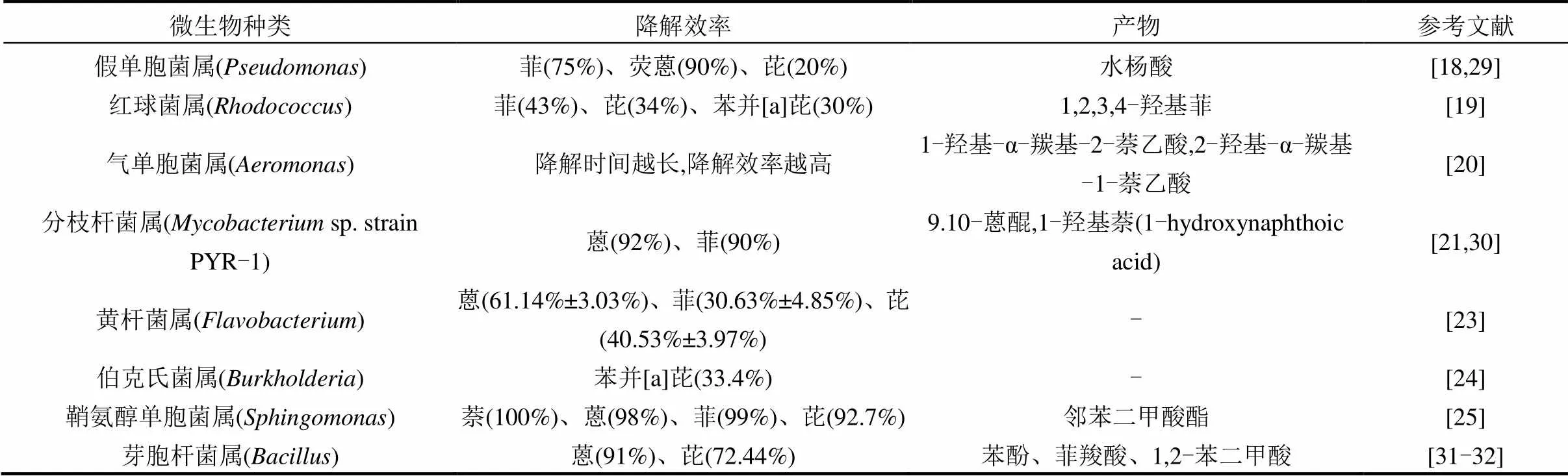
表1 基于纯培养方法分离的能够降解PAHs的微生物
注:-表示产物暂不清楚.
然而,环境中已发现的微生物99%以上都无法在实验室纯培养条件下得到.培养基富营养化、微生物生长因子缺乏以及对微生物间复杂联系的忽视和破坏,都是导致传统培养条件未培养微生物无法生长繁殖的主要原因[48].1982年Colwell实验室将这些微生物称作活的但未培养状态[30].未培养微生物分布极其广泛,种类数量丰富多样,且体内含有多种活性化合物和新型基因,在物种进化、自然界新成代谢中扮演重要角色,并成为各个领域极具研究价值的微生物资源[49].尽管分离、纯化培养这些被定义为未培养的微生物存在着困难与挑战,但研究者认为未培养微生物蕴藏的基因、物种、功能多样性信息具有十分重要的研究意义[50].关于未培养微生物的基因、群落结构、功能、遗传进化等方面的研究受到了越来越多人的关注.Kim等[51]研究发现牛瘤胃中未培养细菌种类与可培养细菌种类同样丰富,未培养细菌对牛瘤胃发酵纤维具有重要作用.Ouyang等[52]对3种不同制药废水处理系统的曝气池内污泥微生物群落进行研究,发现未培养的念珠菌、厌氧绳菌和胚泡菌对废水中铵态氮的去除均有贡献.为便于研究人员快速识别分类复杂的未培养微生物,2017年Konstantinidis等[53]提出采用一种新的基于基因组的分类方法,全面描述未培养微生物.研究未培养微生物在环境中的种类、群落结构以及作用机制,有助于探究污染物PAHs在土壤中的原位降解机理,识别受污染土壤中降解PAHs的原位未培养微生物,最大限度的发挥复杂环境中PAHs微生物降解功能.
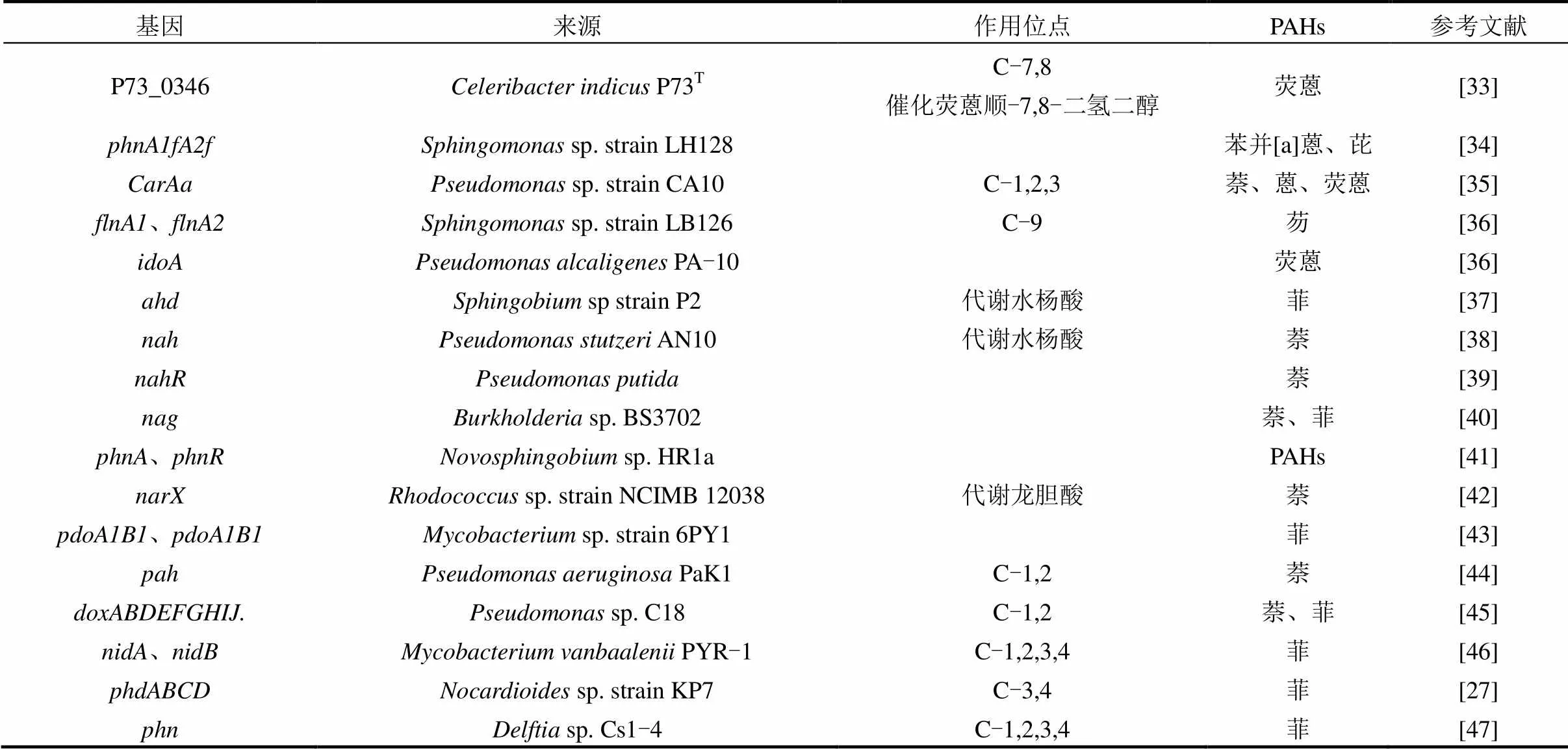
表2 PAHs降解功能基因
鉴于未培养微生物的复杂性,分离、提纯培养分析技术会干扰和破坏原有微生物状态、群落组成与结构,而原位表征未培养微生物技术则更加准确和全面.荧光原位杂交技术、稳定同位素探针技术、宏基因组学技术、转录组学、单细胞技术、磁性纳米材料技术等基于非培养手段的研究方法,已经被应用于PAHs降解功能微生物的原位识别.非培养手段可以揭示在复杂的生态环境中,有哪些微生物参与污染物降解过程,这些微生物在降解过程中主要做什么,以及它们是如何发挥作用的[54].本文对这6种技术的原理、特点以及应用进行归纳总结,以获得较为全面的、能够解析PAHs降解微生物群落的技术概况,为相关领域研究者提供参考依据.
2 基于非培养手段的PAHs降解微生物的识别
2.1 荧光原位杂交技术(FISH)
荧光原位杂交技术是20世纪80年代末发展起来的一种利用非放射性荧光标记的原位杂交技术.其原理是应用特殊的荧光标记物标记核酸探针,根据碱基互补配对原则,将标记探针与目标序列原位杂交,在荧光显微镜下直接观察,或将杂交后的标记探针与荧光素分子耦联复合物结合,进行相对定性、定位、定量检测目标序列的杂交结果.利用标记探针与特定环境基因组中DNA分子杂交,可以表征目标微生物的丰度以及种群分布.该技术具备高灵敏度,高分辨率,可以同时检测多种核苷酸序列,以及快速、安全、可靠的优势.荧光原位杂交技术或与其他相结合,可以原位表征特定环境中降解PAHs的目标微生物丰度、代谢过程以及种群分布.
Sanches等[55]从炼油化工厂废水中富集厌氧微生物群落,探究其对菲和苊的生物降解能力.利用新一代测序技术和荧光原位降解技术表征降解PAHs的群落由附属于和的细菌组成.Hesham等[56]利用酵母菌去除废水中的PAHs,用活性污泥接种、构建了3个不同的系统,并通过PCR-DGGE技术和FISH技术分析对比3个系统中酵母菌的结构以及丰度.结果表明,去除高分子量PAHs的酵母菌主要是来自其中两种生物增强系统接种的5株酵母菌,这两种接种了外来酵母菌的生物增强系统对高分子量PAHs的降解具有显著作用.
2.2 稳定同位素探针技术(SIP)
稳定同位素探针技术(SIP)是一种基于同位素示踪方法鉴别复杂群落中参与某种物质代谢的功能微生物技术,能够将微生物群落结构与其功能相耦合.SIP方法的工作原理如图1所示(以DNA-SIP为例).向环境样品中原位添加同位素标记(13C、15N、18O等)的化合物,经过一段时间的培养,样品中的功能微生物同化标记化合物为磷脂脂肪酸(PLFA)[57]、DNA[58]、RNA[59]、蛋白质[60]等生物标记物.分离微生物中的生物标记物分子并对其中的重同位素进行下游分析,即可获得环境功能微生物信息,揭示其在环境化合物转化中的群落以及功能.
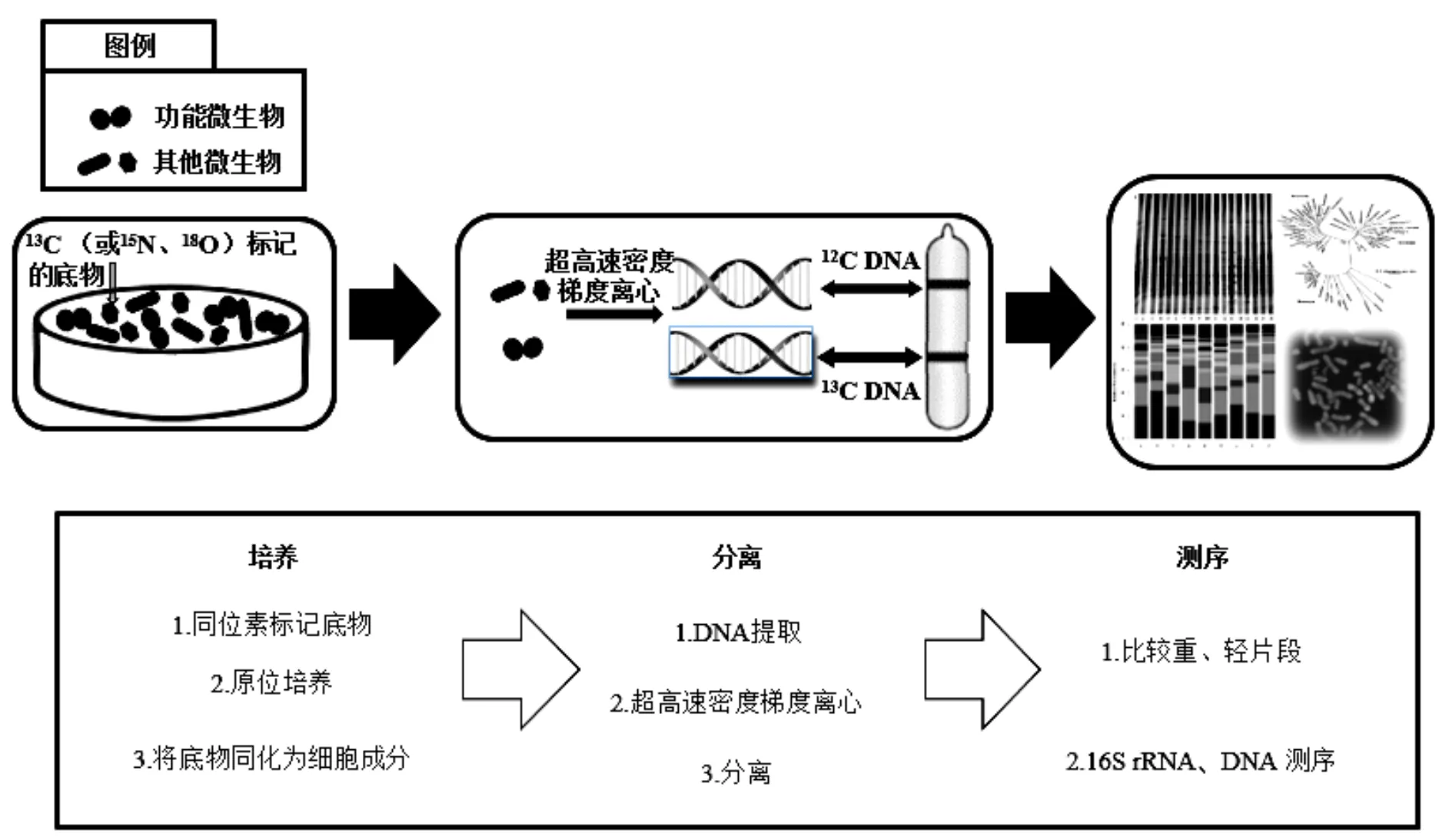
图1 DNA-SIP技术路线
1998年,PLFA-SIP技术首次应用于研究甲烷氧化细菌[61].但当时人们缺乏对目标分子的充分了解,且PLFA-SIP技术分辨率低,导致构建分类信息十分困难.到2000年,研究者成功利用DNA-SIP技术从含有13C-甲醇的土壤中,辨认出-和在甲醇原位代谢过程中发挥主要作用[62].目前,SIP技术主要以DNA[58]、RNA[59]和蛋白质[60]作为生物标志物.其中,DNA分子比RNA更稳定,作为一种信息量最为丰富的生物标记物,DNA分子有最高的分类学分辨率,应用最为广泛.但是微生物基因组中的GC含量会增加未标记DNA的浮力密度,影响标记DNA的有效分离[63-64],且需要较长的培养时间.相比之下,RNA标记独立于细胞分裂,这使得RNA-SIP技术更灵敏.但从环境样品中获取的mRNA浓度低,离心不完全[65],分类精度低且不稳定,以及cRNA缺乏完善的功能基因表达数据库[63]等因素,限制了RNA-SIP技术的发展.蛋白质-SIP技术也独立于细胞分裂,具有灵敏度高、培养时间短以及快速标记等特点,但是相比于其他标记技术,蛋白质-SIP技术的分类精度更低[63].
SIP技术与其他技术结合,已经广泛应用于探究土壤、水、沉积物等环境介质中参与生化反应的功能微生物群落特征,包括废水处理中硝化氧化微生物功能的识别[66-68],海洋微生物的遗传特性及代谢机制研究[69-71],植物根际微生物与C元素循环关系解析[59,72],以及土壤环境中降解功能微生物的识别等[73-74].应用SIP技术识别PAHs降解功能微生物的报道如表3所示.可以看出,目前的研究大多以13C标记PAHs底物,采用DNA-SIP技术研究不同种类PAHs(萘、蒽、菲、荧蒽、芘等)的降解功能微生物.研究的环境介质包括森林土壤、沉积物以及受污染土壤等,由于其含有不同的矿物质、有机物以及腐殖质,具有完全不同的理化性质,因此发掘的微生物数量以及种类也各不相同. Song等[74]的研究,首次利用DNA-SIP技术发现森林土壤中相关细菌直接参与荧蒽的代谢. Rochman等[75]利用SIP技术,在加拿大油砂尾矿库表面检测到家族和是降解萘的主要微生物.SIP方法实现了单一微生物向复杂微生物群落研究的转变,为在群落整体水平,系统研究微生物在自然环境中重要生理过程的分子调控机制,定向发掘重要微生物资源和生物技术开发提供了关键技术支撑.
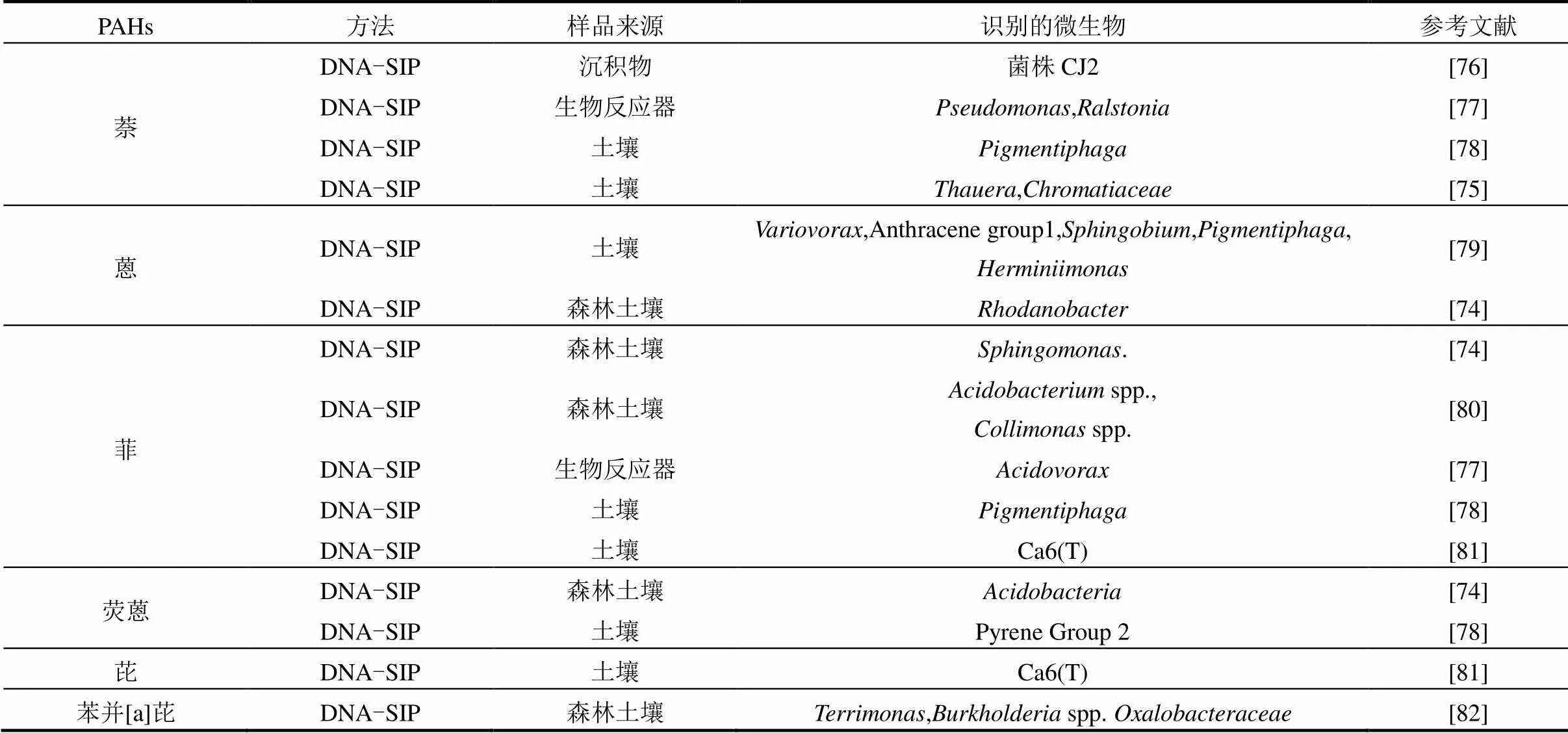
表3 SIP技术在降解PAHs中的应用
2.3 基于高通量测序的宏基因组学技术(Metagenomics)
20世纪80年代基因组学问世,涵盖基因组学、转录组学、蛋白质组学及代谢组学的生物组学内容逐渐丰富.1998年Handelsman等[83]第一次提出宏基因组学一词.不同于其他生物组学,宏基因组学直接从环境样品中提取所有微生物的总DNA进行研究,避开传统分离、培养微生物技术的缺点和困难,全面分析特定环境中微生物群落的组成多样性、遗传多样性、演替与进化,探究全部微生物的种间联系以及与环境的相互作用.宏基因组学技术包括环境总DNA提取,宏基因组文库的构建、筛选、测序以及分析(如图2所示).
利用直接在土壤中裂解微生物细胞提取DNA或先分离细胞再提取DNA的方法,提取土壤中总微生物的DNA.根据所获得DNA选择合适的载体和宿主细胞,将DNA片段连接到载体上并导入宿主菌,构建宏基因组文库.对大量复杂的宏基因组文库筛选有利于快速鉴定功能基因和生物活性物质.宏基因文库的筛选分为序列驱动型筛选和功能驱动型筛选.序列驱动型筛选利用已知基因作为分子探针通过PCR或分子杂交技术筛选克隆基因,以获取某一类结构或功能相似的分子和基因.功能驱动型筛选则利用某种特殊手段的目标活性物质,检测其表达基因,发现新的生物活性物质.相比于功能驱动型筛选,序列驱动型筛选效率更高,但是其依赖于已知基因而不能发现新基因序列[84].
高测序通量及测序速度、低测序成本与污染以及生物信息技术的优化推动着宏基因组学测序技术不断进步[85-86].其中,基于高通量检测技术的宏基因组学测序技术有2种,分别为高通量测序技术和基因芯片技术.基因芯片技术是利用一块固定含有大量已知序列分子探针的小型芯片进行杂交测序,具有高微生物检测深度、高通量性和定量性,但是基因芯片技术只能获取样品中已知物种基因信息而不能检测出新基因或新物种[87].相比之下,高通量测序技术应用更广泛,它以罗氏454测序技术和Illumina测序技术为代表,也被称作第二代测序技术,具有准确性高,信息识别真实可靠以及筛选新基因,发现新物种的特性;但其发现群落中丰度较低的微生物能力仍待提高[64,87-88].将测序所得数据进行存储和序列分析处理,通过比对、提炼信息,分析微生物的群落组成、结构以及生态影响.

图2 宏基因组学技术原理[86]
随着与高通量测序技术应运而生的国际宏基因组大数据库和分析平台的建立和完善,使得宏基因组学技术的应用更为广泛[89].其对微生物多样性的探索遍及海洋[90]、森林[91]、极端环境[92]、气候变化[93]、农业食品[94]、临床医学[95]、环境污染修复[96]等各个领域.尤其是将宏基因组学技术应用到土壤、沉积物等复杂微生物环境中,揭示其中蕴藏的巨大基因、物种、群落资源,为环境修复提供理论依据.2014年,Mason等[97]采集墨西哥湾原油泄漏点2010年9月~10月期间的深海1500m处表层沉积物,提取样品DNA构建16S rRNA宏基因组文库,并利用Illumina技术测序.最终再现了样品碳氢化合物的降解途径以及特定化合物的降解基因,发现了PAHs对表层沉积物中的微生物群落结构有消极影响. Zafra等[98]向PAHs污染土壤中接种能够降解PAHs的微生物群落,利用宏基因组学技术发现接种降解微生物群落能够显著提高土著微生物对PAHs的降解能力,降解基因丰度也发生明显变化.Zhao等[99]从北京周边的一个焦化厂土壤中采样,利用高通量测序手段,探究了其中荧蒽的降解微生物群落、功能基因以及代谢产物,与其它技术结合发现该焦化厂土壤中的和相似丰度高于先前研究的受碳氢化合物污染的土壤中的相似丰度,它们对原位降解PAHs发挥重要作用,其研究结果为复杂微生物群落构建了第一个PAHs降解的协同代谢网.
2.4 转录组学技术
中心法则阐述遗传信息由DNA经过转录传递给RNA,但这并不意味着DNA分子上的所有基因都会被转录,Hangauer等[100]研究证明有85%以上的基因会进行转录,而剩下不到15%的基因都不会参与表达.因此,基于RNA分子的研究在揭示基因表达水平、调控规律以及功能基因方面相比DNA分子研究更有优势.随着基因组学的建立,以某一状态下特定组织或细胞转录出的RNA分子为研究对象的转录组学随后诞生.其研究范围不仅包括能够编码蛋白的mRNA也包括非编码rRNA、tRNA等.目前,无法对RNA直接测序,所以将RNA转化为DNA并对DNA测序分析是转录组学技术的主要思路.相比于微阵列测序技术对物种特性探测存在的偏差,基于高通量测序的RNA测序技术(RNA-Seq)具有更高的灵敏度和精度,承担了转录组学技术的大部分测序任务[101],也是转录组学的研究热点.利用RNA-Seq技术不仅可以在全基因组的尺度上描述编码RNAs和非编码RNAs的转录差异,还有助于探究整个转录水平的基因表达与生物进化的内在联系,识别不同生态环境下的基因转录表达差异以及基因对环境变化的适应潜力[101].
RNA-Seq技术包括实验设计、RNA分离、文库准备、高通量测序以及生物信息学分析几个阶段.其中,mRNA分离、富集以及测序深度、文库大小是关系RNA-Seq技术成功进行的主要因素[102].细胞中的总RNA分子90%以上由rRNA分子组成, mRNA分子只占1%~2%[102],而这极少的mRNA功能分子才是我们的研究重点.目前,研究人员常用Oligo(dT)磁珠提取生物组织或细胞中的mRNA[103-104].再将mRNA片段化之后在引物作用下反转录为cDNA第一链、第二链等,之后扩增cDNA获得cDNA文库,利用高通量技术测序并对数据结果进行分析,即可得到样品的基因表达水平.
转录组学技术广泛应用于临床医药、生态环境、资源开发利用、健康风险评估等领域[101,105-106].对环境生态领域的探究在土壤[107]、水体[108]、极端环境[109]、气候变化[110]等多方面均有涉足.利用转录组学技术,对比基因表达水平在不同生态环境下的动态变化,能够揭示环境胁迫对动植物或微生物体内基因表达的影响.Singh等[107]从转录水平上论述了土壤中的重金属对植物的胁迫作用.他们认为土壤中重金属胁迫会诱导大量的基因以及蛋白质来连接传递信号,这些基因中有一部分是负责编码转录因子的调控基因,转录因子则负责在机体受到胁迫时调控各种反应基因;另一部分功能基因则负责编码代谢各种化合物的酶,这些基因共同操纵着植物在重金属胁迫下的耐受性.在PAHs污染的土壤中,微生物为应对环境因子的变化其基因表达也会受到影响,这种响应可能体现在单个微生物个体的变化、微生物个体与个体间的变化、整个微生物群落结构或功能的变化.目前,转录组学技术对PAHs的研究主要集中在对其生态毒性的探讨以及对人体的健康风险评估[111-113],较少关注土壤中PAHs的生物降解过程.这可能是由于土壤相比于海洋、湖泊等其他介质成分更为复杂,提取生物的mRNA更困难,且mRNA分子的稳定性低于DNA分子.尽管如此,有研究者在几年前就试图从转录组学的角度阐明土壤中PAHs对微生物的基因表达的影响以及污染物代谢途径.2012年Alexandre等[114]向低PAHs背景值的土壤中加入污染物菲,探究土壤样品中的PAHs对土著微生物基因表达的影响.他们从菲完全去除的土样中提取和分离mRNA、反转录为cDNA并进行测序分析.结果显示出与芳香类化合物代谢、呼吸作用以及应激反应相关的基因转录本都有不同程度的增加,与PAHs降解相关的微生物和数量增加.这是第一次利用转录学技术研究土壤微生物群落对污染物的原位响应,它成功的将PAHs的代谢与相应微生物联系起来,从基因表达的层面揭示微生物代谢PAHs的机理.这也表明,连接基因表达与微生物功能桥梁的转录组学技术在探究PAHs等污染物质原位降解机制的巨大潜力,有助于深入探究微生物群落与功能的内在联系.2016年,Meckenstock等[115]应用转录组学联合其他生物化学方法揭示萘的生物降解途径,这是第一次在生物化学和基因水平阐述萘的厌氧降解途径,极大促进了PAHs厌氧代谢以及代谢微生物的生态学研究.尽管土壤的复杂性给转录组学的应用带来阻碍,但其中包涵丰富的微生物及功能基因资源,借助于转录组学对基因转录—表达功能进行深入探究,使得原位识别功能微生物群落结构方面更准确,也为土壤修复工程提供理论指导.
2.5 单细胞技术(Sing-cell technology)
尽管宏基因组学技术和转录组学技术能够分别在DNA分子和基因表达与调控的层面上揭示功能微生物在复杂生态环境下的响应机制,但这两种技术在发现新功能基因和物种丰度极小的未培养微生物方面仍有局限性[59],甚至不能提供明确的基因信息而模糊了特定微生物的代谢过程以及进化历程[116],单细胞技术的兴起弥补了宏基因组学技术以及其他组学技术在微生物领域的认识不足.
来源相同的细胞在分化过程中也会体现出不同细胞间的异质性,生物学家们认为生物学的最基本问题之一就是探究基因调控机制在细胞分化以及组织灵活、协调工作中是如何起作用的[117],而单细胞技术则是解决这一问题的新方法.它可以从微生物细胞中提供基因信息,确定在环境生物地球化学循环中占主导地位的未培养微生物的代谢潜力,开发细胞尺度上定量描述细胞特性的方法.如Rinke等[118]利用单细胞技术发现古生菌中有一种与肽聚糖合成有关的酶,由于古生菌中不含肽聚糖,他们推测这种酶可能是古生菌与其他细胞相互作用的结果.
单细胞基因组技术流程如图3所示.包括单细胞的获取、细胞裂解、全基因组扩增、基因测序与数据分析.获取单细胞的方式最常用的新方法是基于流式细胞术的荧光激活细胞分选法(FACS)和微流控芯片技术.其中,FACS具有高通量、高灵活性、高纯度的优点被广泛使用,但是FACS分离法对细胞存活率有一定影响[119];液滴微流控芯片技术需要样品数量极少,价格低廉,污染小且高度自动化、高通量[120].目前实现单细胞全基因组扩增最新的方法是多重置换扩增技术(MDA).Li等[121]实现了在微流控芯片上将MDA技术与芯片外PCR技术相结合,成功发现了细胞的异质性.单细胞基因组技术现阶段面临的主要挑战是将定量、综合的基因组学与微观的分辨率结合,提高从组织到细胞的分析精度[117].单个细胞的基因组与转录组存在一定的内在联系,而且在单个细胞内同时测量多种分子类型比在多个细胞内测量多种分子类型更有优势,所以Macaulay等[122]提出在单个细胞中应用多组学分析,有助于明确基因型与表现型的关系,同时了解从DNA到RNA再到蛋白质以及细胞表型的调控机制.利用基因组序列构建细胞谱系图,同时利用转录组序列反映细胞的类型和状态,增强对细胞异质性以及种群结构、进化的理解.
2009年,Stepanauskas[116]课题组建立了第一个专门从事单细胞基因组学研究的毕格罗实验室中心(SCGC),他们处理的微生物细胞来源涉及土壤、海洋、地下深层以及其他环境.单细胞技术的应用十分广泛,它可以从细胞水平阐述环境中PAHs等污染物的刺激对微生物细胞结构、形态的影响以及微生物间复杂、微妙的相互作用.Chen等[123]利用流式细胞术探究电子垃圾场中的苯并[a]芘对降解微生物的细胞膜电位、细胞周期、细胞凋亡以及蛋白质表达的影响.结果显示,苯[a]芘是影响细胞膜电位以及破坏膜结构的主要原因.此外,在污染物的胁迫下,微生物为适应不良环境其参与污染物转运与代谢的酶以及蛋白质表达有显著变化.细胞膜作为细胞与污染物直接接触的一道屏障,在接触过程中发生着形态、结构的改变:PAHs等污染物通过与细胞膜接触,改变、破坏膜的脂质结构,导致细胞膜空隙增大、细胞渗透性增强[124],这种改变为细胞内化污染物提供条件.虽然单细胞技术在探究土壤中PAHs生物降解的应用较少,但该技术在研究功能微生物群落的进化、结构和功能方面,具有广阔的前景.
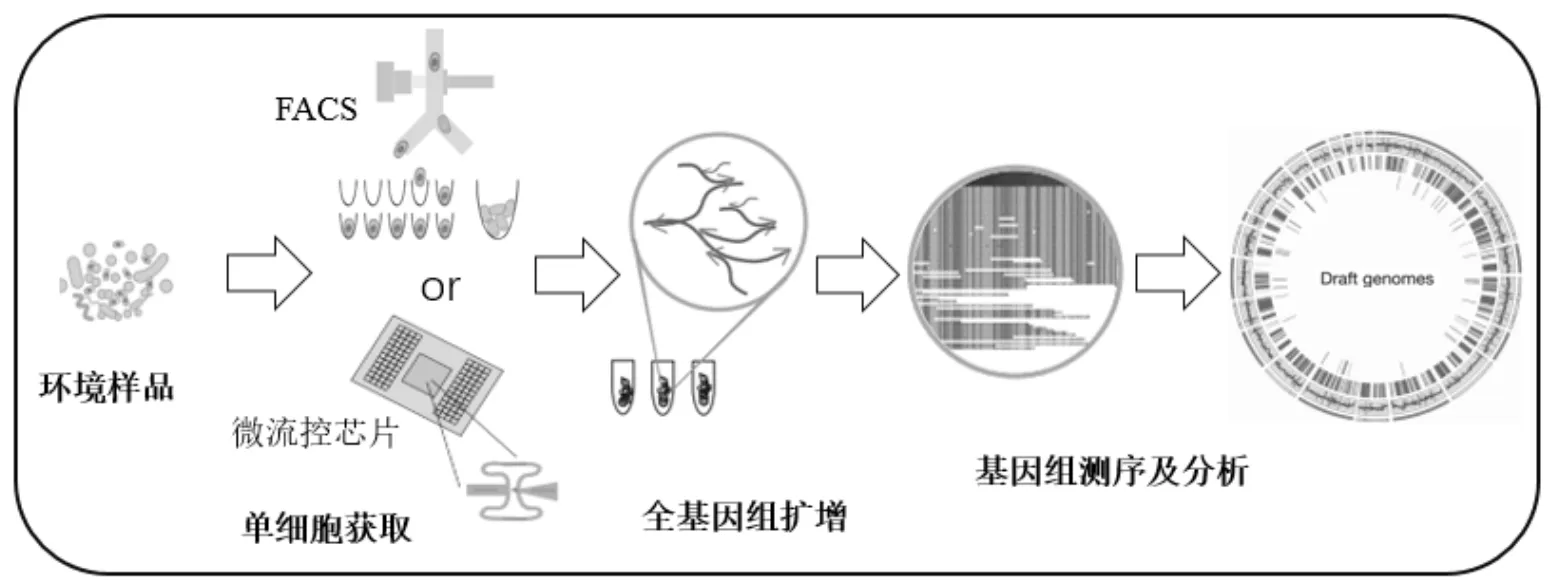
图3 单细胞基因组技术流程图[118]
2.6 磁性纳米材料技术
磁性纳米材料(MMI)是一种利用磁性纳米材料从复杂微生物群落中分离活的功能微生物的新型技术,该技术的最大优势在于不依赖于对底物碳、氮源的标记,能够从复杂环境中分离活的功能微生物,这对准确认识功能微生物的生理生态特性和功能具有重要意义.
MMI技术流程示意如图4所示.在一个复杂的微生物系统中,用磁性纳米材料(MNPs)功能化系统中的所有细胞;向其中加入底物(如PAHs等),系统中能够代谢底物为自身生长、繁殖提供物质能源的磁性功能化细胞将会连续大量分裂、繁殖;分裂的细胞磁性会逐渐被稀释直到完全失去磁性,而系统中另一部分不能代谢底物或代谢较慢的微生物细胞分裂、繁殖速率较慢,仍保留磁性;在外加磁场的作用下,系统内带有磁性的细胞和不带磁性的细胞实现分离;收集得到活的功能微生物[125].
MNPs在物理、化学、生物各学科都有应用研究[126-128],其优点是尺寸小、有磁性且反应活性高.在环境学领域的应用主要集中在基于磁性纳米材料高效分离、提取细胞内核酸[128-131].利用MNPs分离复杂群落中的活细胞,前提是需要将MNPs有效吸附在细胞表面且保证细胞的活性与正常生理功能.Zhang等[132]的研究表明,纳米颗粒与细胞的接触、摄入过程中会发生物理、化学作用,其中纳米颗粒的大小、形状以及物化性质都会影响这个过程,甚至产生毒性.2011年Zhang等[133]研发了一种具有生物相容性的MNPs功能化细胞的方法.该方法将聚烯丙基胺盐酸盐固定化的MNPs反复分离和纯化,获得具有良好生物相容性和细胞友好性的MNPs.利用这种方法功能化的细胞带有磁性且功能化效率达到99.96%.2015年,Zhang等[125]利用这种方法,研究焦化厂废水处理污泥中的微生物,发现未培养的spp.是负责原位降解苯酚的主要微生物,这是第一次将MMI应用于原位识别污泥中的未培养功能微生物,并利用Biolog平板研究其对碳源和氮源的利用情况.2016年Wang等[134]将MMI技术应用到原位识别受原油污染土壤中降解正烷烃的微生物,识别出和是降解正烷烃的主要微生物.

图4 MMI技术流程(改自文献[125])
3 前景与展望
环境中存在着大量数量庞大、功能多样的未培养微生物,然而人们对它们的种类以及在地球生物化学循环中的作用还知之甚少.目前对PAHs降解微生物的研究还处于实验室纯培养阶段,但是复杂介质中未培养微生物对PAHs的降解未忽视.FISH、SIP、宏基因组学、转录组学、单细胞以及磁性纳米材料技术,从基因和细胞的不同层次,提供了不依赖于纯培养方法的识别PAHs降解微生物的解决途径.目前,DNA-SIP技术已经较为广泛地应用于对降解PAHs功能微生物的原位识别,其它技术也有少量应用在污染物生物降解领域.虽然这些非培养手段避开了传统培养方法的局限性,为我们原位了解未培养微生物基因多样性、群落多样性、遗传多样性提供了可能性,但是在现有研究水平下这些技术仍有不足.如DNA-SIP技术依赖于底物的同位素标记,组学技术容易忽略丰度低的微生物,磁性纳米材料技术依赖于功能微生物在研究时间内的大量分裂等.
为了更全面认识未培养微生物的基因表达、遗传进化、群落结构与功能,笔者认为可以从以下几方面完善非培养手段的不足.
3.1 多组学技术相结合.宏基因组学技术从DNA分子水平识别环境中未培养微生物,转录组学技术则从转录水平探究,单细胞技术更进一步探究基因表达与功能的内在联系.将三者结合起来,同时进行多组学分析则可以将遗传信息在微生物体内的所有参与过程整合起来,避免对微生物认识的片面化.
3.2 高效利用最新测序技术和分析方法,加强数据存储、分析平台的建设.近几年来,测序成本不断降低,读取长度逐渐增加,高效利用最新的测序技术和分析手段会提高我们的数据精确度.同时,大量的数据信息以及数据源的共享要求我们必须加强信息平台的建设.
3.3 对萌芽技术(MMI技术)深入探究验证.MMI技术目前仍不够成熟,对识别分裂能力较差的功能细胞能力有限,有待探究完善.
[1] Tarafdar A, Sinha A. Public health risk assessment with bioaccessibility considerations for soil PAHs at oil refinery vicinity areas in India [J]. The Science of the Total Environment, 2017,616- 617:1477-1484.
[2] Yin W, Hou J, Xu T, et al. Obesity mediated the association of exposure to polycyclic aromatic hydrocarbon with risk of cardiovascular events [J]. The Science of the Total Environment, 2018,616-617:841-854.
[3] Tavera B I, Tames F, Silva J A, et al. Biomonitoring levels and trends of PAHs and synthetic musks associated with land use in urban environments [J]. The Science of the Total Environment, 2017,618: 93-100.
[4] Tu Y T, Ou J H, Tsang D C W, et al. Source identification and ecological impact evaluation of PAHs in urban river sediments: A case study in Taiwan [J]. Chemosphere, 2018,194:666-674.
[5] Yadav I C, Devi N L, Li J, et al. Polycyclic aromatic hydrocarbons in house dust and surface soil in major urban regions of Nepal: Implication on source apportionment and toxicological effect [J]. The Science of the Total Environment, 2017,616-617:223-235.
[6] 康 杰,胡 健,朱兆洲,等.太湖及周边河流表层沉积物中PAHs的分布、来源与风险评价[J]. 中国环境科学, 2017,37(3):1162-1170.
[7] Wang C L, Zou X Q, Zhao Y F, et al. Distribution pattern and mass budget of sedimentary polycyclic aromatic hydrocarbons in shelf areas of the Eastern China Marginal Seas [J]. Journal of Geophysical Research-Oceans, 2017,122(6):4990-5004.
[8] Xiao Y H, Tong F C, Kuang Y W, et al. Distribution and Source Apportionment of Polycyclic Aromatic Hydrocarbons (PAHs) in Forest Soils from Urban to Rural Areas in the Pearl River Delta of Southern China [J]. International Journal of Environmental Research and Public Health, 2014,11(3):2642-2656.
[9] Wang Y, Zhang S, Cui W, et al. Polycyclic aromatic hydrocarbons and organochlorine pesticides in surface water from the Yongding River basin, China: Seasonal distribution, source apportionment, and potential risk assessment [J]. The Science of the Total Environment, 2018,618:419-429.
[10] Tong R P, Yang X Y, Su H R, et al. Levels, sources and probabilistic health risks of polycyclic aromatic hydrocarbons in the agricultural soils from sites neighboring suburban industries in Shanghai [J]. The Science of the Total Environment, 2018,616-617:1365-1373.
[11] Kuppusamy S, Thavamani P, Venkateswarlu K, et al. Remediation approaches for polycyclic aromatic hydrocarbons (PAHs) contaminated soils: Technological constraints, emerging trends and future directions [J]. Chemosphere, 2017,168:944-968.
[12] Peris P M, Gómez J C, Herbert J H, et al. Ecological restoration of a former gravel pit contaminated by a massive petroleum sulfonate spill. A case study: Arganda del Rey. Madrid (Spain) [J]. Ecological Engineering, 2017,100:73-88.
[13] Lin W, Guo C, Zhang H, et al. Electrokinetic-Enhanced Remediation of Phenanthrene-Contaminated Soil Combined withsp GY2B and Biosurfactant [J]. Applied Biochemistry and Biotechnology, 2016,178(7):1325-1338.
[14] Srivastava V J, Hudson J M, Cassidy D P.solidification andchemical oxidation combined in a single application to reduce contaminant mass and leachability in soil [J]. Journal of Environmental Chemical Engineering, 2016,4(3):2857-2864.
[15] Cheng M, Zeng G, Huang D, et al. Hydroxyl radicals based advanced oxidation processes (AOPs) for remediation of soils contaminated with organic compounds: A review [J]. Chemical Engineering Journal, 2016, 284:582-598.
[16] Chen M, Xu P, Zeng G, et al. Bioremediation of soils contaminated with polycyclic aromatic hydrocarbons, petroleum, pesticides, chlorophenols and heavy metals by composting: Applications, microbes and future research needs [J]. Biotechnology Advances, 2015,33(6):745-755.
[17] Liu S H, Zeng G M, Niu Q Y, et al. Bioremediation mechanisms of combined pollution of PAHs and heavy metals by bacteria and fungi: A mini review [J]. Bioresource Technology, 2017,224:25-33.
[18] Lin M, Hu X, Chen W, et al. Biodegradation of phenanthrene bysp. BZ-3, isolated from crude oil contaminated soil [J]. International Biodeterioration and Biodegradation, 2014,94:176-181.
[19] Song X, Xu Y, Li G, et al. Isolation, characterization ofsp. P14capable of degrading high-molecular-weight polycyclic aromatic hydrocarbons and aliphatic hydrocarbons [J]. Marine Pollution Bulletin, 2011,62(10):2122-2128.
[20] Nie M, Nie H, Cao W, et al. Phenanthrene Metabolites from a New Polycyclic Aromatic Hydrocarbon-Degrading Bacteriumsubsp.Strain NY4 [J]. Polycyclic Aromatic Compounds, 2016,36(2):132-151.
[21] Moody J D, Freeman J P, Doerge D R, et al. Degradation of phenanthrene and anthracene by cell suspensions ofsp. strain PYR-1 [J]. Applied and Environmental Microbiology, 2001, 67(4):1476-1483.
[22] Chikere C B, Chikere B O, Okpokwasili G C. Bioreactor-based bioremediation of hydrocarbon-polluted Niger Delta marine sediment, Nigeria [J]. Biotech., 2012,2(1):53-66.
[23] Ni N, Song Y, Shi R, et al. Biochar reduces the bioaccumulation of PAHs from soil to carrot (Daucus carota L.) in the rhizosphere: A mechanism study [J]. The Science of the Total Environment, 2017,601:1015-1023.
[24] Song M, Luo C, Jiang L, et al. Identification of Benzo a pyrene-Metabolizing Bacteria in Forest Soils by Using DNA-Based Stable-Isotope Probing [J]. Applied and Environmental Microbiology, 2015,81(21):7368-7376.
[25] Hesham A E, Mawad A M M., Mostafa Y M, et al. Biodegradation Ability and Catabolic Genes of Petroleum-DegradingStrain ASU-06Isolated from Egyptian Oily Soil [J]. Biomed Research International, 2014,2014(1):127674.
[26] Seo J S, Keum Y S, Hu Y, et al. Phenanthrene degradation insp P1-1: Initial 1,2-, 3,4- and 9,10-dioxygenation, and meta- and ortho-cleavages of naphthalene-1,2-diol after its formation from naphthalene-1,2-dicarboxylic acid and hydroxyl naphthoic acids [J]. Chemosphere, 2006,65(11):2388-2394.
[27] Saito A, Iwabuchi T, Harayama S. Characterization of genes for enzymes involved in the phenanthrene degradation insp. KP7 [J]. Chemosphere, 1999,38(6):1331-1337.
[28] Yen K M, Gunsalus I C. Regulation of naphthalene catabolic genes of plasmid NAH7 [J]. Journal of Bacteriology, 1985,162(3):1008-1013.
[29] Chebbi A, Hentati D, Zaghden H, et al. Polycyclic aromatic hydrocarbon degradation and biosurfactant production by a newly isolatedsp. strain from used motor oil-contaminated soil [J]. International Biodeterioration and Biodegradation, 2017,122: 128-140.
[30] Xu H S. Survival and viability of nonculturableEscherichia coli andVibrio cholerae in the estuarine and marine environment [J]. Microbial Ecology, 1982,8(4):313-323.
[31] Tarafdar A, Sinha A, Masto R E. Biodegradation of anthracene by a newly isolated bacterial strain,AT.ISM.1, isolated from a fly ash deposition site [J]. Letters in Applied Microbiology, 2017,65(4):327-334.
[32] Meena S S, Sharma R S, Gupta P, et al. Isolation and identification ofYB3 from an effluent contaminated site efficiently degrades pyrene [J]. Journal of Basic Microbiol., 2016, 56(4):369-378.
[33] Cao J, Lai Q, Yuan J, et al. Genomic and metabolic analysis of fluoranthene degradation pathway inP73(T) [J]. Scientific Reports, 2015,5:7741.
[34] Schuler L, Jouanneau Y, Chadhain S M N, et al. Characterization of a ring-hydroxylating dioxygenase from phenanthrene-degradingsp. strain LH128able to oxidize benz a anthracene [J]. Applied Microbiology and Biotechnology, 2009,83(3):465-475.
[35] Nojiri H, Nam J W, Kosaka M, et al. Diverse oxygenations catalyzed by carbazole 1,9a-dioxygenase fromsp. Strain CA10 [J]. Journal of Bacteriology, 1999,181(10):3105-3113.
[36] Schuler L, Chadhain S M N, Jouanneau Y, et al. Characterization of a novel angular dioxygenase from fluorene-degradingsp. strain LB126 [J]. Applied and Environmental Microbiology, 2008, 74(4):1050-1057.
[37] Pinyakong O, Habe H, Yoshida T, et al. Identification of three novel salicylate 1-hydroxylases involved in the phenanthrene degradation ofsp. strain P2 [J]. Biochemical and Biophysical Research Communications, 2003,301(2):350-357.
[38] Bosch R, Garcíavaldés E, Moore E R. Complete nucleotide sequence and evolutionary significance of a chromosomally encoded naphthalene-degradation lower pathway fromAN10 [J]. Gene, 2000,245(1):65-74.
[39] Park H H, Lim W K, Shin H J. In vitro binding of purified NahR regulatory protein with promoter[J]. Biochimica Et Biophysica Acta-General Subjects, 2005,1725(2):247-255.
[40] Izmalkova T Y, Sazonova O I, Kosheleva I A, et al. Phylogenetic Analysis of the Genes for Naphthalene and Phenanthrene Degradation insp. Strains [J]. Russian Journal of Genetics., 2013, 49(6):609-616.
[41] Segura A, Hernándezsánchez V, Marqués S, et al. Insights in the regulation of the degradation of PAHs insp. HR1a and utilization of this regulatory system as a tool for the detection of PAHs [J]. The Science of the Total Environment., 2017,590:381-393.
[42] Liu T T, Xu Y, Liu H, et al. Functional characterization of a gene cluster involved in gentisate catabolism insp. strain NCIMB 12038 [J]. Applied Microbiology and Biotechnology, 2011,90(2):671-678.
[43] Krivobok S, Kuony S, Meyer C, et al. Identification of pyrene- induced proteins insp. strain 6PY1: Evidence for two ring-hydroxylating dioxygenases [J]. Journal of Bacteriology, 2003, 185(13):3828-3841.
[44] Takizawa N, Iida T, Sawada T, et al. Nucleotide sequences and characterization of genes encoding naphthalene upper pathway ofPaK1 andOUS82 [J]. Journal of Bioscience and Bioengineering, 1999,87(6):721-731.
[45] Denome S A, Stanley D C, Olson E S, et al. Metabolism of dibenzothiophene and naphthalene instrains: complete DNA sequence of an upper naphthalene catabolic pathway [J]. Journal of Bacteriology, 1993,175(21):6890-6901.
[46] Stingley R L, Khan A A, Cerniglia C E. Molecular characterization of a phenanthrene degradation pathway inPYR-1 [J]. Biochemical and Biophysical Research Communications, 2004,322(1):133-146.
[47] Hickey W J, Chen S, Zhao J. Theisland: a new genomic island encoding catabolism of polynuclear aromatic hydrocarbons [J]. Frontiers in Microbiology, 2012:3(125):1-12.
[48] 张作艳,原 野,丁立建,等.未培养微生物原位培养技术研究进展[J]. 天然产物研究与开发, 2018,38(5):907-913.
[49] Pulschen A A, Bendia A G, Fricker A D, et al. Isolation of Uncultured Bacteria from Antarctica Using Long Incubation Periods and Low Nutritional Media [J]. Frontiers in Microbiology, 2017,8(12):1346.
[50] 崔晓龙,徐丽华,文孟良,等.未培养微生物资源[J]. 微生物学通报, 2005,(3):144-146.
[51] Kim M, Yu Z. Quantitative comparisons of select cultured and uncultured microbial populations in the rumen of cattle fed different diets [J]. Journal of Animal Science and Biotechnology, 2012,3(4): 193-198.
[52] Ouyang E, Liu Y, Ouyang J, et al. Effects of different wastewater characteristics and treatment techniques on the bacterial community structure in three pharmaceutical wastewater treatment systems [J]. Environmental Technology, 2017,(11):1-13.
[53] Konstantinidis K T, Rosselló-Móra R, Amann R. Uncultivated microbes in need of their own taxonomy [J]. ISME Journal, 2017, 11(11):2399-2406.
[54] Neufeld J D, Wagner M, Murrell J C. Who eats what, where and when? Isotope-labelling experiments are coming of age [J]. Isme Journal, 2007,1(2):103.
[55] Sanches S, Martins M, Silva A F, et al. Bioremoval of priority polycyclic aromatic hydrocarbons by a microbial community with high sorption ability [J]. Environmental Science and Pollution Research International, 2017,24(4):3550-3561.
[56] Hesham E L, Khan S, Yu T, et al. Biodegradation of high molecular weight PAHs using isolated yeast mixtures: application of meta- genomic methods for community structure analyses [J]. Environmental Science and Pollution Research International., 2012,19(8):3568-3578.
[57] Johnsen A R, Winding A, Karlson U, et al. Linking of microorganisms to phenanthrene metabolism in soil by analysis of C-13-labeled cell lipids [J]. Applied and Environmental Microbiology, 2002,68(12): 6106-6113.
[58] Coyotzi S, Pratscher J, Murrell J C, et al. Targeted metagenomics of active microbial populations with stable-isotope probing [J]. Current Opinion in Biotechnology, 2016,41:1-8.
[59] Gkarmiri K, Mahmood S, Ekblad A, et al. Identifying the Active Microbiome Associated with Roots and Rhizosphere Soil of Oilseed Rape [J]. Applied and Environmental Microbiology, 2017,83(22): AME.01938-17.
[60] Jehmlich N, Vogt C, Lünsmann V, et al. Protein-SIP in environmental studies [J]. Current Opinion in Biotechnology, 2016,41:26-33.
[61] Boschker H T S, Nold S C, Wellsbury P, et al. Direct linking of microbial populations to specific biogeochemical processes by13C-labelling of biomarkers [J]. Nature, 1998,392(6678):801-805.
[62] Radajewski S, Ineson P, Parekh N R, et al. Stable-isotope probing as a tool in microbial ecology [J]. Nature, 2000,403(6770):646-649.
[63] Lueders T, Dumont M G, Bradford L, et al. RNA-stable isotope probing: from carbon flow within key microbiota to targeted transcriptomes [J]. Current Opinion in Biotechnology, 2016,41:83-89.
[64] Youngblut N D, Buckley D H. Intra-genomic variation in G plus C content and its implications for DNA stable isotope probing [J]. Environmental Microbiology Reports, 2014,6(6):767-775.
[65] Gao J, Pan K, Li H, et al. Application of GelGreen (TM) in Cesium Chloride Density Gradients for DNA -Stable Isotope Probing Experiments [J]. PLos One, 2017,12(1):1-13.
[66] Martinezcruz K, Leewis M C, Herriott I C, et al. Anaerobic oxidation of methane by aerobic methanotrophs in sub-Arctic lake sediments [J]. The Science of the Total Environment, 2017,607-608:23-31.
[67] Srithep P, Pornkulwat P, Limpiyakorn T. Contribution of ammonia-oxidizing archaea and ammonia-oxidizing bacteria to ammonia oxidation in two nitrifying reactors [J]. Environmental Science and Pollution Research International, 2018,25(3):1-12.
[68] Ziels R M, Sousa D Z, Stensel H D, et al. DNA-SIP based genome- centric metagenomics identifies key long-chain fatty acid-degrading populations in anaerobic digesters with different feeding frequencies [J]. ISME Journal, 2018,12(1):112-123.
[69] Cunliffe M, Hollingsworth A, Bain C, et al. Algal polysaccharide utilisation by saprotrophic planktonic marine fungi [J]. Fungal Ecology, 2017,30:135-138.
[70] Bryson S, Zhou L, Chavez F, et al. Phylogenetically conserved resource partitioning in the coastal microbial loop [J]. ISME Journal, 2017,11(12):2781-2792.
[71] Orsi W D, Wilken S, Campo J D, et al. Identifying protist consumers of photosynthetic picoeukaryotes in the surface ocean using stable isotope probing [J]. Environmental Microbiology, 2018,20(2):815- 827.
[72] Costa D P D, Dias A C F, Cotta S R, et al. Changes of bacterial communities in the rhizosphere of sugarcane under elevated concentration of atmospheric CO2[J]. Global Change Biology Bioenergy, 2018,10(2):137-145.
[73] Dai S, Liu Q, Zhao J, et al. Ecological niche differentiation of ammonia-oxidising archaea and bacteria in acidic soils due to land use change [J]. Soil Research, 2018,56(1):71-79.
[74] Song M, Jiang L, Zhang D, et al. Bacteria capable of degrading anthracene, phenanthrene, and fluoranthene as revealed by DNA based stable-isotope probing in a forest soil [J]. Journal of Hazardous Materials, 2016,308:50-57.
[75] Rochman F F, Sheremet A, Tamas I, et al. Benzene and Naphthalene Degrading Bacterial Communities in an Oil Sands Tailings Pond [J]. Front Microbiol., 2017,8(12):1845.
[76] Jeon C O, Park W, Padmanabhan P, et al. Discovery of a bacterium, with distinctive dioxygenase, that is responsible for in situ biodegradation in contaminated sediment [J]. Proceedings of the National Academy of Sciences of the United States of America, 2003,100(23):13591-13596.
[77] Singleton D R, Powell S N, Sangaiah R, et al. Stable-isotope probing of bacteria capable of degrading salicylate, naphthalene, or phenanthrene in a Bioreactor treating contaminated soil [J]. Applied and Environmental Microbiology, 2005,71(3):1202-1209.
[78] Jones M D, Crandell D W, Singleton D R, et al. Stable-isotope probing of the polycyclic aromatic hydrocarbon-degrading bacterial guild in a contaminated soil [J]. Environmental Microbiology, 2011, 13(10):2623-2632.
[79] Jones M D, Singleton D R, Sun W, et al. Multiple DNA Extractions Coupled with Stable-Isotope Probing of Anthracene-Degrading Bacteria in Contaminated Soil [J]. Applied and Environmental Microbiology, 2011,77(9):2984-2991.
[80] Jiang L, Song M, Luo C, et al. Novel Phenanthrene-Degrading Bacteria Identified by DNA-Stable Isotope Probing [J]. Plos One, 2015,10(6):1-14.
[81] Corteselli E M, Aitken M D, Singleton D R.gen. nov., sp nov., a bacterium within the family, isolated from contaminated soil, capable of degrading aromatic compounds [J]. International Journal of Systematic and Evolutionary Microbiology, 2017,67(2):311-318.
[82] Song M, Luo C, Jiang L, et al. Identification of Benzo a pyrene- Metabolizing Bacteria in Forest Soils by Using DNA-Based Stable- Isotope Probing [J]. Applied and Environmental Microbiology, 2015, 81(21):7368-7376.
[83] Handelsman J, Rondon M R, Brady S F, et al. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products [J]. Chemistry and Biology, 1998,5(10):245-249.
[84] Montella S, Amore A, Faraco V. Metagenomics for the development of new biocatalysts to advance lignocellulose saccharification for bioeconomic development [J]. Critical Reviews in Biotechnology, 2016,36(6):998-1009.
[85] Kim M, Lee K H, Yoon S W, et al. Analytical tools and databases for metagenomics in the next-generation sequencing era [J]. Genomics and Informatics, 2013,11(3):102-113.
[86] Neelakanta G, Sultana H. The use of metagenomic approaches to analyze changes in microbial communities [J]. Microbiology Insights, 2013,6:37-48.
[87] Zhou J, He Z, Yang Y, et al. High-Throughput Metagenomic Technologies for Complex Microbial Community Analysis: Open and Closed Formats [J]. Microbiology, 2015,6(1):1-17.
[88] Lindgreen S, Adair K L, Gardner P P. An evaluation of the accuracy and speed of metagenome analysis tools [J]. Scientific Reports, 2016,6:1-14.
[89] Klemetsen T, Raknes I A, Fu J, et al. The MAR databases: development and implementation of databases specific for marine metagenomics [J]. Nucleic Acids Research, 2018,46(D1):D692-D699.
[90] Smith T E, Pond C D, Pierce E, et al. Accessing chemical diversity from the uncultivated symbionts of small marine animals [J]. Nature Chemical Biology, 2018,14(2):179-185.
[91] Mendes L W, Tsai S M. Distinct taxonomic and functional composition of soil microbiomes along the gradient forest-restinga- mangrove in southeastern Brazil [J]. Antonie Van Leeuwenhoek International Journal of General and Molecular Microbiology, 2018, 111(1):101-114.
[92] Becraft E D, Dodsworth J A, Murugapiran S K, et al. Genomic Comparison of Two Family-Level Groups of the Uncultivated NAG1Archaeal Lineage from Chemically and Geographically Disparate Hot Springs [J]. Frontiers in Microbiology, 2017,8(10):1-10.
[93] Ms. Sylwia Zielińska, Dr. Dorota Kidawa, Prof. Lech Stempniewicz, et al. Environmental DNA as a valuable and unique source of information about ecological networks in Arctic terrestrial ecosystems [J]. Environmental Reviews, 2017,25(3):282-291.
[94] Sarkar S F, Poon J S, Lepage E, et al. Enabling a sustainable and prosperous future through science and innovation in the bioeconomy at Agriculture and Agri-Food Canada [J]. New Biotechnology, 2018,40: 70-75.
[95] Douglas G M, Hansen R, Cma J, et al. Multi-omics differentially classify disease state and treatment outcome in pediatric Crohn's disease [J]. Microbiome, 2018,6(13):1-12.
[96] Sahu N, Deori M, Ghosh I. Metagenomics Study of Contaminated Sediments from the Yamuna River at Kalindi Kunj, Delhi, India [J]. Genome Announcements, 2018,6(1):1-7.
[97] Mason O U, Scott N M, Gonzalez A, et al. Metagenomics reveals sediment microbial community response to Deepwater Horizon oil spill [J]. Isme Journal Multidisciplinary Journal of Microbial Ecology, 2014,8(7):1464-1475.
[98] Zafra G, Taylor T D, Absalón A E, et al. Comparative metagenomic analysis of PAH degradation in soil by a mixed microbial consortium [J]. Journal of Hazardous Materials, 2016,318:702-710.
[99] Zhao J K, Li X M, Ai G M, et al. Reconstruction of metabolic networks in a fluoranthene-degrading enrichments from polycyclic aromatic hydrocarbon polluted soil [J]. Journal of Hazardous Materials, 2016,318:90-98.
[100]Hangauer M J, Vaughn I W, Mcmanus M T. Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs [J]. Plos Genetics, 2013,9(6): e1003569.
[101]Todd E V, Black M A, Gemmell N J. The power and promise of RNA-seq in ecology and evolution [J]. Molecular Ecology, 2016, 25(6):1224-1241.
[102]Conesa A, Madrigal P, Tarazona S, et al. A survey of best practices for RNA-seq data analysis [J]. Genome Biology, 2016,17(1):13.
[103]Haile S, Corbett R D, Macleod T, et al. Increasing quality, throughput and speed of sample preparation for strand-specific messenger RNA sequencing [J]. BMC Genomics, 2017,18(1):515.
[104]Kaluzna M, Kuras A, Mikicinski A, et al. Evaluation of different RNA extraction methods for high-quality total RNA and mRNA from[J]. European Journal of Plant Pathology, 2016,146(4):893-899.
[105]Molyneaux P L, Willisowen S A G, Cox M J, et al. Host-Microbial Interactions in Idiopathic Pulmonary Fibrosis [J]. American Journal of Respiratory and Critical Care Medicine, 2017,195(12):1640-1650.
[106]Gruzieva O, Xu C J, Breton C V, et al. Epigenome-Wide Meta- Analysis of Methylation in Children Related to Prenatal NO2Air Pollution Exposure [J]. Environmental Health Perspectives, 2017, 125(1):104-110.
[107]Singh S, Parihar P, Singh R, et al. Heavy Metal Tolerance in Plants: Role of Transcriptomics, Proteomics, Metabolomics, and lonomics [J]. Frontiers in Plant Science, 2016,6(1143):1-36.
[108]Evans T G, Padillagamiño J L, Kelly M W, et al. Ocean acidification research in the 'post-genomic' era: Roadmaps from the purple sea urchin[J]. Comparative Biochemistry and Physiology a-Molecular and Integrative Physiology, 2015,185: 33-42.
[109]Goltsman D S A, Comolli L R, Thomas B C, et al. Community transcriptomics reveals unexpected high microbial diversity in acidophilic biofilm communities [J]. The ISME journal, 2015,9(4): 1014-1023.
[110]Imadi S R, Kazi A G, Ahanger M A, et al. Plant transcriptomics and responses to environmental stress: an overview [J]. Journal of Genetics, 2015,94(3):525-537.
[111]Brinkmann M, Koglin S, Eisner B, et al. Characterisation of transcriptional responses to dioxins and dioxin-like contaminants in roach (Rutilus rutilus) using whole transcriptome analysis [J]. The Science of the Total Environment, 2016,541:412-423.
[112]Curtis L R, Bravo C F, Bayne C J, et al. Transcriptional changes in innate immunity genes in head kidneys from-challenged rainbow trout fed a mixture of polycyclic aromatic hydrocarbons [J]. Ecotoxicology and Environmental Safety, 2017,142:157-163.
[113]Labib S, Williams A, Kuo B, et al. A framework for the use of single-chemical transcriptomics data in predicting the hazards associated with complex mixtures of polycyclic aromatic hydrocarbons [J]. Archives of Toxicology, 2017,91(7):2599-2616.
[114]De Menezes Alexandre, Clipson N, Doyle E. Comparative metatranscriptomics reveals widespread community responses during phenanthrene degradation in soil [J]. Environmental Microbiology, 2012,14(9): 2577-2588.
[115]Meckenstock R U, Boll M, Mouttaki H, et al. Anaerobic Degradation of Benzene and Polycyclic Aromatic Hydrocarbons [J]. J. Journal of Molecular Microbiology and Biotechnology, 2016,26(1-3):92-118.
[116]Stepanauskas R. Single cell genomics: an individual look at microbes [J]. Current Opinion in Microbiology, 2012,15(5):613-620.
[117]Tanay A, Regev A. Scaling single-cell genomics from phenomenology to mechanism [J]. Nature, 2017,541(7637):331-338.
[118]Rinke C, Schwientek P, Sczyrba A, et al. Insights into the phylogeny and coding potential of microbial dark matter [J]. Nature, 2013,499(7459):431-437.
[119]Gross A, Schoendube J, Zimmermann S, et al. Technologies for Single-Cell Isolation [J]. International Journal of Molecular Sciences, 2015,16(8):16897-16919.
[120]Shembekar N, Chaipan C, Utharala R, et al. Droplet-based microfluidics in drug discovery, transcriptomics and high-throughput molecular genetics [J]. Lab on a Chip, 2016,16(8):1314-1331.
[121]Li R, Zhou M, Yue C, et al. Multiple single cell screening and DNA MDA amplification chip for oncogenic mutation profiling [J]. Lab on a Chip, 2018,18(5):723-734.
[122]Macaulay I C, Ponting C P, Voet T. Single-Cell Multiomics: Multiple Measurements from Single Cells [J]. Trends in Genetics, 2017, 33(2):155-168.
[123]Chen S, Yin H, Chang J, et al. Physiology and bioprocess of single cell ofin bioremediation of co-existed benzo a pyrene and copper [J]. Journal of Hazardous Materials, 2017,321:9-17.
[124]Croxton A N, Wikfors G H. The use of flow cytometric applications to measure the effects of PAHs on growth, membrane integrity, and relative lipid content of the benthic diatom,[J]. Marine Pollution Bulletin, 2015,91(1):160-165.
[125]Zhang D, Berry J P, Zhu D, et al. Magnetic nanoparticle-mediated isolation of functional bacteria in a complex microbial community [J]. Isme Journal, 2015,9(3):603-614.
[126]Ghahremani M, Aslani A, Hosseinnia M, et al. Direct and indirect measurement of the magnetocaloric effect in bulk and nanostructured Ni-Mn-In Heusler alloy [J]. AIP Advances, 2018,8(5):056426.
[127]Tzitzios V K. Chemical synthesis of L10Fe-Pt-Ni alloy nanoparticles [J]. AIP Advances, 2018,8(5):056210.
[128]Paul T, Basu S, Sarkar K. SPION-mediated soil DNA extraction and comparative analysis with conventional and commercial kit-based protocol [J]. 3Biotech, 2014,4(6):669-677.
[129]Kamat V, Pandey S, Paknikar K, et al. A facile one-step method for cell lysis and DNA extraction of waterborne pathogens using a microchip [J]. Biosensors and Bioelectronics, 2018,99:62-69.
[130]Wang J, Ali Z, Si J, et al. Simultaneous Extraction of DNA and RNA from Hepatocellular Carcinoma (Hep G2) Based on Silica-Coated Magnetic Nanoparticles [J]. Journal of Nanoscience and Nanotechnology, 2017,17(1):802-806.
[131]Tang Y, Zou J, Chao M, et al. Highly Sensitive and Rapid Detection ofBased on Magnetic Enrichment and Magnetic Separation [J]. Theranostics, 2013,3(2):85-92.
[132]Zhang S, Gao H, Bao G. Physical Principles of Nanoparticle Cellular Endocytosis [J]. Acs Nano, 2015,9(9):8655-8671.
[133]Zhang D, Fakhrullin R F, Ozmen M, et al. Functionalization of whole-cell bacterial reporters with magnetic nanoparticles [J]. Microbial Biotechnology, 2011,4(1):89-97.
[134]Wang X, Zhao X, Li H, et al. Separating and characterizing functional alkane degraders from crude-oil-contaminated sites via magnetic nanoparticle-mediated isolation [J]. Research in Microbiology, 2016,167(9/10):731-744.
致谢:感谢中国博士后科学基金(2017M620626)和中央高校基本科研业务费(FRF-TP-16-063A1)对本研究的支持.
Identification and characterization on PAHs-degrading microorganisms via cultivation-independent approaches.
CHEN Ya-ting, JIANG Bo*, XING Yi*, ZHANG Na-na, LIAN Lu-ning, LU Pei
(Beijing Key Laboratory of Resource-oriented Treatment of Industrial Pollutants, School of Energy and Environmental Engineering, University of Science & Technology Beijing, Beijing 100083, China)., 2018,38(9):3562~3575
This review discussed the cultivation-independent approaches which have been developed to identify functional-yet- uncultivated soil microbesand revealed their functions in PAHs degradation. PAHs-degrading microbes, degradation mechanisms and functional genes were summarized. The principles, characteristics and applications of cultivation-independent methods, including Fluorescencehybridization (FISH), Stable-isotope probing (SIP), Metagenomics, Transcriptomics, Single-cell technology and Magnetic nanoparticle-Mediated Isolation technology (MMI) were also discussed. Examples were given on the application of these cultivation-independent approaches in identifying PAHs-degrading microbes. SIP technology is the most stable and widely used in the study of PAHs degradation mechanism. As a novel cultivation-independent approach, MMI technology provides a deep insight into exploring functional-yet-uncultivated PAHs-degrading microbes, which stands out for its ability to separate active functional microbes.
functional-yet-uncultivated microbes;PAHs biodegradation;soils;functional microorganisms;cultivation-independent approaches
X53
A
1000-6923(2018)09-3562-14
陈亚婷(1994-),女,新疆伊犁人,北京科技大学硕士研究生,主要研究污染场地的微生物修复.发表论文2篇.
2018-03-09
中国博士后科学基金资助项目(2017M620626);中央高校基本科研业务费资助项目(FRF-TP-16-063A1)
* 责任作者, 姜博, 讲师, jiangbo_seee@ustb.edu.cn; 邢奕, 教授, xingyi@ustb.edu.cn
