Taxonomy, Morphology and Phylogeny of a New Oligotrich Ciliate–Omegastrombidium hongkongense n. sp. (Protozoa: Ciliophora) from Clear Water Bay, Hong Kong
2018-08-24SHENZhuoLIUWeiweiZHANGShuwenYIZhenzhenandLIUHongbin
SHEN Zhuo, LIU Weiwei, ZHANG Shuwen, YI Zhenzhen, and LIU Hongbin,
Taxonomy, Morphology and Phylogeny of a New Oligotrich Ciliaten. sp. (Protozoa: Ciliophora) from Clear Water Bay, Hong Kong
SHEN Zhuo1), 2),*, LIU Weiwei3), ZHANG Shuwen2), YI Zhenzhen4), and LIU Hongbin2), *
1),,,519082,2),,,,3),,,510301,4),,,510631,
One new marine oligotrich ciliate,n. sp., was isolated from a bloom ofnear Port Shelter, Hong Kong. The morphology and infraciliature of this new species were studied on both living and protargol-stained specimens. Its phylogenetic position was discussed based on the sequence of the small subunit rRNA gene.is different from its congeners with special characters. The cells are usually heart-shaped, and the cell size usuallyis (20–35)×(20–30)μm.Its deep buccal cavity extends obliquely to about 1/2 of cell length.It shows prominent apical protrusion.The adoral zone of membranelles is divided into 17–19 collar membranelles and four buccal membranelles. It has one ball-like macronucleus.The girdle kinety forms a closed loop which obliquely surrounds the body.The ventral kinety and thigmotactic membranelles are not observed. The SSU rRNA sequence ofwas close to those ofandwith approximately 99% similarity. In the phylogenetic trees,can be grouped withandspecies with very low support (16% ML).
infraciliature; marine ciliate; morphology; Oligotrichia;
1 Introduction
Marine planktonic ciliates are diverse and play an important role in marine ecosystem as components of the marine energy flux (Pierce and Turner, 1992; Jiang., 2011; Chen., 2015). The oligotrich genuswas separated from genusby Agatha (2004a). According to Agatha (2004a, 2011), the most distinctive differences betweenand other genera in Strombidiidae are (i) the pattern of girdle kinety that horizontally orientated on dorsal side while kinety ends extend to posterior end of ventral side; and (ii) the oral primordium anterior to the horizontal girdle kinety in division cell. Two species,and,were identified fromand grouped intoaccording to the characters mentioned above..(Florentin, 1901) Agatha, 2004 and(Alekperov, 1985) Agatha, 2004 are the only two species inso far.
With surveys of ciliates diversity in Port Shelter of Hong Kong, many oligotrich ciliates were sampled. According to the detailed observations of both living and silver-stained specimens,as well as the comparison with known species, we concluded that this is a new member of. The phylogenetic analyses were conducted based on SSU rRNA sequences.The detailed results are presented and discussed here.
2 Materials and Methods
2.1 Sample Collection, Isolation and Identification
n. sp. was collected from(red) bloom with sampling water directly in Port Shelter, Kowloon, Hong Kong (114˚16´E, 22˚16´N) on December 18, 2014 when the water temperature was about 24℃, salinity was 32.5 and pH was 8.2 (Figs.1a–b).
crawled on the surface ofwith the ventral side facing down, and usually aggregated around clusters ofprogametes and feed on them.was immediately isolated from cell surface ofwith micropipette under anatomical lens. No cultures were established in the laboratory. Living cells were observed by bright field and differential interference contrast microscopy. Protargol staining was used to reveal the infraciliature (Wilbert, 1975). Illustrations of live specimens were based on direct observations and light micrographs, while those of protargol-impregnated specimens were made with the help of a camera (Spot insight QE, USA) at 1000× magnification. Counts and measurements on stained specimens were performed at a magnification of 1000 (Liu., 2015b; Song., 2015b). Terminology and classification wereconductedaccording to Agatha (2004a, 2014) and Lynn, (2008) respectively.
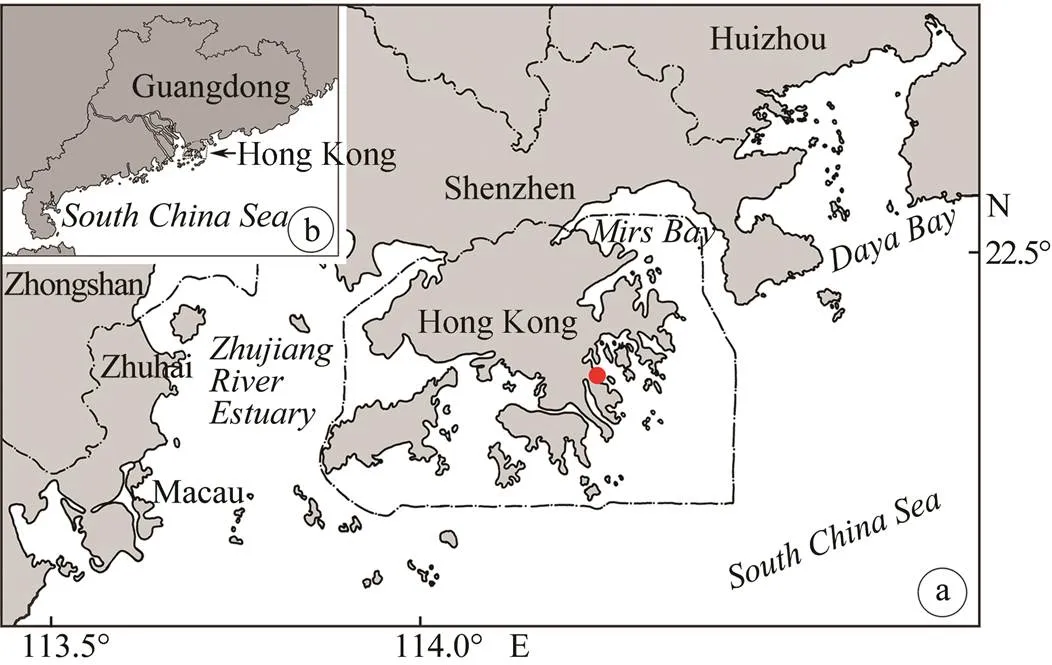
Fig.1 Sampling site. (a) Hong Kong area, red dot shows sampling site; (b) Guangdong Province, indicates the relative location of Hong Kong.
2.2 SSU rRNA Extraction, PCR Amplificationand Sequencing
Genomic SSU rRNA ofn. sp. was extracted using REDExtract-N-Amp™ Tissue PCR Kit (Sigma, St. Louis, USA) according to Liu. (2015a). The PCR amplification of SSU rRNA was performed with primers Euk A (5’-AAC CTG GTT GAT CCT GCC AGT-3’) and Euk B (5’-TGA TCC TTC TGC AGG TTC ACC TAC-3’) (Medlin., 1988). Genomic SSU rRNA extraction, Cloning, PCR amplification, and sequencing ofn. sp. were performed following the methods of Yi. (2014). The SSU rRNA sequence has been deposited in GenBank with accession no. KT730064.
2.3 Phylogenetic Analysis
The SSU rRNA sequence ofn. sp. and those of 77 ciliates from GenBank database (for accession numbers see Fig.4) were used for the construction of phylogenetic tree with the methods reported by Song. (2015b).was used as the out-group taxon. MrModeltest v.2.0 (Nylander 2004) was implemented to select the GTR+I+G evolutionary model as the best model with Akaike Information Criterion, which was used for Bayesian inference (BI) analysis. The BI analysis was performed with MrBayes 3.1.2 (Ronquist and Huelsenbeck 2003), which was run for 1000000 generations with a sample frequency of 100 generations. The first 2500 trees were discarded as burn-in.
Maximum likelihood (ML) analysis, using the GTR plus INVGAMMA substitution model, was performed by RaxML-HPC2 (Stamatakis., 2008) at the CIPRES website (http://www.phylo.org/). Searches for the best tree were conducted starting from 100 random trees, and 1000 bootstrap replicates were performed with the multi- parametric algorithm.
A maximum parsimony (MP) tree was computed in PAUP* 4.0 (Swofford, 2002). Parameters are as following: heuristic search, 10 random-addition sequences and 1000 bootstrap replicates.
3 Results
3.1 Diagnosis
Smallabout 30×25μmwith heart-shaped cell, slightly flattened dorsoventrally, staining cell about 30×30μm; deep and prominent buccal cavity extending obliquely to about 1/2 of cell length; prominent apical protrusion; the adoral zone of membranelles divided into 17–19 collar membranelles and four buccal membranelles, no thigmotactic membranelle; the girdle kinety, with 12–15 dikinetids, forming a closed loop which located higher on left than right side of the body; ventral kinety absent; extrusome acicular shaped, around 8×0.5μm, arranged in a single row under the girdle kinety; one ball-shaped macronucleus (Table 1).

Table 1 Morphometric characters of O. hongkongense n. sp. from protargol impregnated specimens
Notes: All data based on protargol impregnated and randomly selected specimens. Measurements in μm. GK, girdle kinety; Mean, arithmetic mean; n, number of specimens investigated; SD, standard deviation.
3.2 Typelocality and Host
Type locality: coastal waters off Clear Water Bay (Hong Kong, 114˚16´E, 22˚16´N). Host:(Ehrenberg) Macartney, 1810. Host tissue: external.
3.3 Type Specimens
One holotype and one paratype slides of protargol-im- pregnated specimens are deposited in Laboratory of Protozoology, Institute of Evolution and Marine Biodiversity, Ocean University of China, Qingdao, China (registration numbers SZ2014-1218-01-3, SZ2014-1218-4)
3.4 Etymology
The Latin word ‘’ refers to the sampling site (Hong Kong) of this species.
3.5 Morphological Description
Cell size mostly (20–35)× (20–30)μm, (25–40)×(25–30)μm after protargol impregnation. Cell usually heart-shaped, slightly flattened dorsoventrally, thickness: width about 2.5:1 (Figs.2a; 3a, b). Anterior end domed forming an oblique hyaline apical protrusion (about 3μm high) at the right side of peristome that can be recognizedbut undetectable after protargol impregnation (Figs.2c; 3a). Buccal cavity deep and prominent, extending obliquely to right and terminating at about equatorial region (Figs.2c; 3a). Pellicle thin, no hemitheca observed. Cytoplasm colorless, often filled with numerous granules(diameter 1–7μm); several food vacuoles (diameter about 5μm) with progametes ofin it, giving cell light grey to light brown appearance (Figs.2a, c). Extrusomes conspicuous, acicular shaped, around 8μm×0.5μm; their attachment sites arranged in a single row under the girdle kinety (Figs.2e; 3b). Single macronucleus, containing manysmall chromatin aggregates (diameter about 1–2μm; Figs.2f; 3f). Micronucleus, contractile vacuole and cytopyge not observed.
Usually the cells crawl on, using the collar membranelles and buccal membranelles for attachment with the ventral side down. Sometimes, they swim in narrow spirals by rotation about the main axis for a little while (Figs.2b; 3c).

Fig.2 O. hongkongense n. sp. from life (a–e), after protargol impregnation (f–l). a, View of a representative individual. b, An O. hongkongense cell on the surface of Noctiluca scintillans, arrow marks O. hongkongense. c, Ventral view of a typical individual, arrow indicates buccal membrane, arrowhead shows apical protrusion. d, Dorsal view, arrow marks food vacuole, arrowheads indicate food particles. e, Dorsal view, arrows show extrusomes. f, Infraciliature, arrows indicate collar membranelles, arrowhead marks endoral membrane. g, Dorsal view, arrow shows the fiber connection between dikinetids of GK, arrowheads indicate girdle kinety. h, Ventral view, arrowhead marks fiber. i, Ventral view, arrowhead shows endoral membrane. j, Early division stage, arrow shows opisthe’s oral primordium, arrowheads mark GK. k, Middle division stage, arrow shows opisthe’s oral primordium, arrowheads mark GK. l, Late division stage, arrow shows opisthe’s oral primordium, arrowheads mark GK. Ma=macronucleus; BM=Buccal membranelles; GK=Girdle kinety. Scale bars=15μm (a, c–e), 10μm (f–i), 500μm (b).
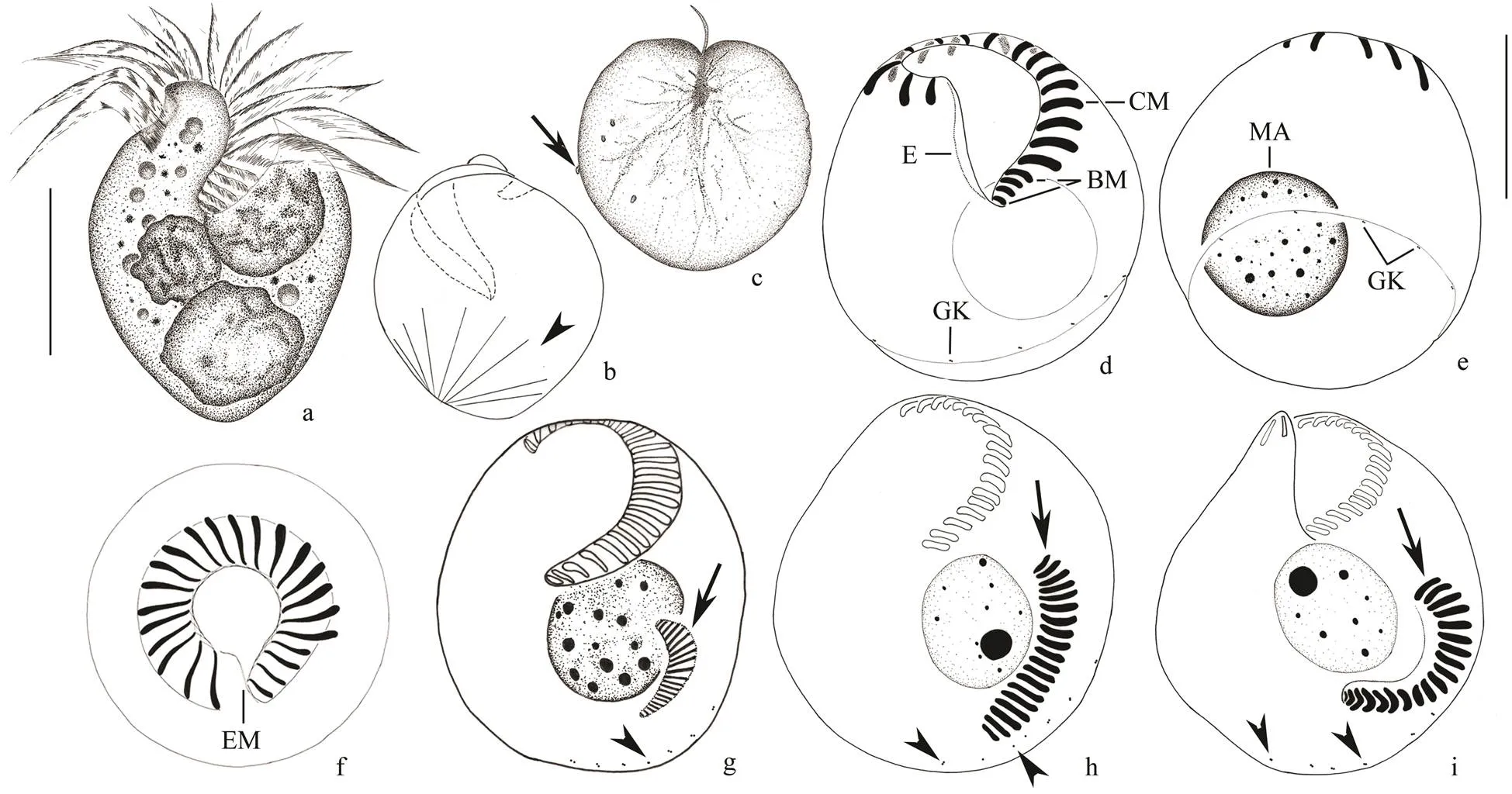
Fig.3 O. hongkongense n. sp. from living samples (a–c) and the samples stained with protargol (d–i). a, Ventral viewof a typical cell. b, Dorsal view, shows extrusomes (arrow). c, O. hongkongense (arrow) on the Noctiluca scuntillans cell. d, Oral apparatus. e–f, Ventral (e) and dorsal (f) views of the same specimen showing the ciliary pattern and nuclear apparatus. g, Early division stage. h, Middle division stage, arrow shows opisthe’s oral primordium, arrowheads mark GK. i, Late division stage, arrow shows opisthe’s oral primordium, arrowheads mark GK; CM, collar membranelles; GK, girdle kinety; BM, buccal membranelles; E, endoral membrane; Ma, macronucleus. Scale bars=10μm (a, c–f).
3.6 Somatic Ciliature
The adoral zone of membranelles divided into 17–19 collar membranelles around the apical protrusion and four buccal membranelles without thigmotactic membranelle. The collar membranelles continued with collar membranelles (Figs.2f, h; 3e), and fiber system under the pellicle connect each membranelle (Fig.3h). Bases of collar membranelles are about 5μm long, cilia is approximately 12μm long(Figs.3a, f) and extend radially when swimming. The bases of the buccal membranelles are about 2–3μm long, gradually slightly short- ening from distal to proximal end (Fig.2h). The endoral membranelles are on the inner wall of the buccal cavity, and are often difficult to be recognized in protargol stained specimens (Figs.2f, i; 3e, f). All somatic kineties are composed of dikinetids (Figs.2f, g; 3e, f). Girdle kinety comprising around 12–15 dikinetids are widely spaced and connected by fiber system (Figs.2f, g; 3e, f). Girdle kinety forming a closed circle obliquely surrounds the body. It is located at equatorial region on dorsal side and extends to posterior end on ventral side. It is positioned higher on left than right side of the body (Figs.2g; 3e). The ventral kinety is absent.
Unfortunately, only opisthe’s oral primordium in a few division stages ofn. sp. has been observed clearly. Opisthe’s oral primordium develops on the left of ventral side, and above girdle kinety and extrusome attachment sites (Figs.1g; 2j). Oral primordium develops and forms an inversely and longitudinally orientated L-shape in middle and late divider (Figs.1h, i; 2k, l). Girdle kinety and extrusome attachment sites are always below oral primordium of opisthe on ventral side (Figs.1g–i; 2j–l).
3.7 SSU rRNA Sequence Analysis and Phylogenetic Tree of O. hongkongense
The SSU rRNA sequence ofn. sp. is 1766bp in length. The sequence similarities betweenn. sp. andand some similar species ranged from 90% to 98%.andboth showed the highest similarity (98%) with our new species in SSU rRNA sequence (Table 2).

Table 3 SSU rRNA sequence similarities and the number of unmatched nucleotidesbetween sequenced congeners and O. hongkongense n. sp.
All three phylogenetic methods resulted in similar tree topologies. The ML tree is shown with support values as an example (Fig.4).Lynnellidae is identified from the Oligotrichia clade with variable supports (64% ML, 0.91 BI) trees. The monophyly of the subclass Choreotrichia is confirmed with high support (99% ML, 1.00 BI, 94% MP). The family Strombidiidae is also supported to be monophyletic. Within the family,andcluster together with variable support values (63% ML, 1.00 BI, 81% MP) and then group withwith a very low support (16% ML) (Fig.4).

Fig.4 Maximum likelihood tree inferred from small subunit rRNA sequences indicating phylogenetic positions of O. hongkongense n. sp. (bold typeface, red arrows). Numbers at the nodes represent support values in the following order: Maximum Likelihood (ML) bootstrap values, Maximum Parsimony (MP) bootstrap values, and Bayesian inference (BI) posterior probabilities. Nodes absent from one of the three phylogenies are indicated by a hyphen instead of a support value. The field in green represents the family Strombidiidae. The scale bar indicates the number of substitutions per 10 nucleotides.
4 Discussion
n. sp. is different from most other common strombidiid species by some living behaviors. It was sampled only at the peak of some of(red) bloom.cells always crawl on the surface ofand can not swim freely for a long time. It might feed on gamete ofFurther observation demonstrates that it could be cultured within the lab for two to three days, but less than 24 hours without. There was no evidence thatcan feed on bacteria or phytoplankton from our studies (Zhang., 2017).
The diagnosis ofis that Strombidiidae with girdle kinety horizontally orientate on dorsal side and kinety ends extending to the posterior end of ventral side, which forms the roughly Ω shape (Agatha, 2004a). The distinctive character that the girdle kinety ventral-dorsal obliquely surrounding the body as well as the oral primordium above the girdle kinety on ventral side in this new species is similar with. The main difference between our form and the diagnosis ofis the completely closed posterior end of girdle kinety on ventral side (Song, 2005; Xu., 2006; Agatha, 2011). However, the completion of circle girdle kinety is not stable in family Strombidiidae, such asin which the alternative close or open of girdle kinety commonly occurs. So, according to the morphological and morphogenetic characteristics, we consider the genusto be the most appropriate taxon to accommodate our form.
Up to now, only twospecies,(Florentin, 1901) Agatha, 2004 and(Alekperov, 1985) Agatha, 2004 have been reported. Butclearly differs frommainly in that girdle kinety open on ventral side and form a roughly Ω-shape (. girdle kinety closed in), having 2 thigmotactic membranelles (. absent in) and ventral kinety (. absent in) (Song., 2000; Agatha, 2004b). Whilecan be separated fromby cell size (150×90μm. 30×25μm in), different patterns of girdle kinety (dorsal portion horizontally positioned, ventral potion running across ventral side of cell and terminating on dorsal side. a closed circle in) and numbers of dikinetids in girdle kinety (around 100. 12–15 in) have also been observed.
The most distinctive character ofis its girdle kinety obliquely surrounds the body,., horizontally orientates on dorsal side and extends to posterior end on ventral side.(Lei., 1999) Aga- tha, 2004 has a similar girdle kinety pattern.can be clearly identified by the number of buccal membranelles (7–9. 4 in), the number of thigmotactic membranelles (3. absent in), the number of dikinetids in ventral kinety (8–16. absent in), pattern of girdle kinety (small gap on ventral side. a closed circle obliquely surrounds the body in) and the number of dikinetids in girdle kinety (approximately 50. 12–15 in) (Song., 2000; Agatha, 2004a).
Twociliates lacking ventral kinety have been reported (Agatha, 2004b), includingAgatha and Riedel-Lorje, 1997 and(Leegaard 1915) Kahl, 1932 (Table 3). The large buccal cavity and absence of a ventral kinety inAgatha and Riedel-Lorjé, 1997 also can be observed in. However, two species can be clearly separated mainly according to the number of buccal membranelles (: 17–19.: 4) and the number of dikinetids in girdle kinety (: 108–152.: 12–15) (Agatha and Riedel- Lorjé, 1997).(Leegaard 1915) Kahl, 1932 has been reported in various coastal marine waters (Montagnes., 1988; Jiang., 2011). It has the similar cell size asand also lacks ventral kinety. But it can be separated from our new species by the number of buccal membranelles (9–15. 4in) (Montagnes., 1988).

Table 3 Comparison of morphometric characters of O. hongkongense n. sp., O. elegans,O. elatum, S. paracalkinsi,S. triquetrum and S. acutum
Notes: BM, buccal membranelles; CM, collar membranelles; GK, girdle kinety; VK, ventral kinety; TM, thigmotactic membranelles; ‘–’, character absent.
Considering the absence of a ventral kinety,is also similar to, andandare in the same group in ML tree. However,can be clearly separated from the latter as it has a dorsal gap in the girdle kinety (. forming a closed circle in), and a horizontally oriented girdle kinety (. obliquely in) (Agatha, 2011).
In the phylogenetic trees,is grouped withspecies first, then related toand. These results confirm the classification of our species intoand the morphology similarity amongthese three species. Given that the morphological similarity betweenandspecies,., girdle kinety completely closed and circle, we supposed that the girdle kinety inprobably represents a derived stage during the evolution of girdle kinety fromto. However, the support values of thebranch are too low to draw any conclusions about its systematic position.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31301868), the Hong Kong Research Grants Council (Nos. GRF 661610 and 661912), the Hong Kong Scholar Programme (No. HKSP13SC03), NSFC-Guangdong Province Joint Foundation (No. U1301235) and Natural Science Foundation of Guangdong Pro- vince (No. S2013040013192). We are grateful to Ms. Pui Yin Lee for her logistical support.
Agatha, S., 2004a. Evolution of ciliary patterns in the Oligotrichida (Ciliophora, Spirotricha) and its taxonomic implications., 107: 153-168.
Agatha, S., 2004b. A cladistic approach for the classification of Oligotrichid ciliates (Ciliophora: Spirotricha)., 43: 201-217.
Agatha, S., 2011. Updated hypothesis on the evolution of oligotrichid ciliates (Ciliophora, Spirotricha, Oligotrichida) based on somatic ciliary patterns and ontogenetic data., 47: 51-56.
Agatha, S., 2014. Redescription of(Leegaard, 1915) Kahl, 1932 (Ciliophora, Spirotricha) based on live observation, protargol impregnation, and scanning electron microscopy., 53: 287-294.
Agatha, S., and Riedel-Lorjé, J. C., 1997. Morphology, infraciliature, and ecology of halteriids and strombidiids (Ciliophora, Oligotrichea) from coastal brackish water basins.,148 (4): 445-459.
Alekperov, I. K., 1985. New free-living ciliates from fresh waters of Azerbaijan., 64: 1461-1467 (in Russian with English summary).
Chen, X., Zhao, X. L., Liu, X. H., Warren, A., Zhao, F. Q., and Miao, M., 2015. Sequencing and analysis of the transcriptome from a non-model ciliate:(Protozoa, Ciliophora) using illumina RNA-Seq technology.,6 (5): 373-385, http://dx.doi.org/10.1007/s13238- 015-0147-3.
Jiang, Y., Xu, H. L., Hu, X. Z., Zhu, M. Z., Al-Rasheid, K. A. S., and Warren, A., 2011. An approach to analyzing spatial patterns of planktonic ciliate communities for monitoring water quality in Jiaozhou Bay, northern China., 62: 227-235.
Lei, Y. L., Xu, K. D., and Song, W. B., 1999. Free living ciliates from marine farming ponds. In:.Song, W.,., eds., Qingdao Ocean University Press, Qingdao, 269-295 (in Chinese).
Liu, W. W., Yi, Z. Z., Lin, X. F., Li, J. M., Al-Farraj, S. A., Al- Rasheid, K. A. S., and Song, W. B., 2015a. Morphology and molecular phylogeny of three new oligotrich ciliates (Protozoa, Ciliophora) from the South China Sea., 174: 653-665.
Liu, W. W, Yi, Z. Z., Xu, D., Clamp, C. J., Li, J. Q., Lin, X. F., and Song, W. B., 2015b. Two new genera of planktonic ciliates and insights into the evolution of the family Strombidiidae (Protista, Ciliophora, Oligotrichia)., 10 (6): e0131726, http://dx.doi.org/10.1371/journal.pone.0131726.
Lynn, D. H., 2008.. 3rd edition. Springer, Dordrecht, 605pp.
Medlin, L., Elwood, H. J., Stickel, S., and Sogin, M. L., 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions., 71: 491-499.
Montagnes, D. J. S., Lynn, D. H., Stoecker, D. K., and Small, E. B., 1988. Taxonomic description of one new species and redescription of four species in the family Strombidiiae (Cilio-
phora, Oligotrichida)., 35: 189-197.
Nylander, J. A., 2004. MrModeltest Ver.2. Department of Systematic Zoology, Evolutionary Biology Center, Uppsala Uni- versity.
Pierce, R. W., and Turner, J. T., 1992. Ecology of planktonic ciliates in marine food webs., 6: 139-181.
Ronquist, F., and Huelsenbeck, J. P., 2003. MRBAYES 3: Ba- yesian phylogenetic inference under mixed models., 19: 1572-1574.
Song, W. B., 2005. Taxonomic description of two new marine oligotrichous ciliates (Protozoa, Ciliophora)., 39: 241-252
Song, W. B., Wang, M., and Warren, A., 2000. Redescriptions of three marine ciliates,Florentin, 1901,Claparède & Lachmann, 1859 andLei, Xu & Song, 1999 (Cilio- phora, Oligotrichida)., 36: 327-342.
Song, W., Zhao, X., Liu, W. W., Hu, X. Z., Al-Farrajc, S. A., Al-Rasheid, K. A. S., Song W. B., and Warren, A., 2015b. Biodiversity of oligotrich ciliates in the South China Sea: Description of three newspecies (Protozoa, Ciliophora, Oligotrichia) with phylogenetic analyses., 13: 608-623.
Stamatakis, A., Hoover, P., and Rougemont, J., 2008. A rapid bootstrap algorithm for the RAxML web servers., 57: 758-771.
Swofford, D. L., 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4. Sunderland, MA: Sinauer Associates.
Wilbert, N., 1975. Eine verbesserte Technik der Protargolimprägnation für Ciliaten., 64: 171-179.
Xu, D. P., Song, W. B., Sun, P., and Chen, X. R., 2006. Morphology and infraciliature of the oligotrich ciliate(Yagiu, 1933) Kahl, 1934 (Protozoa, Ciliophora, Oligotrichida) from the intestine of sea urchinAgassiz., 1113: 33-40.
Yi, Z. Z., Strüder-Kypke, M., Hu, X. Z., Lin, X. F., and Song, W. B., 2014. Sampling strategies for improving tree accuracy and phylogenetic analyses: A case study in ciliate protists, with notes on the genus.,71: 142-148.
Zhang, S. W., Chan, K. Y. K., Shen, Z., Cheung, S. Y., Landry, M. R., and Liu, H. B., 2017. A cryptic marine ciliate feeds on progametes of., 168: 1-11.
(Edited by Qiu Yantao)
(Received March 16, 2017; revised April 18, 2017; accepted October 22, 2017)
© Ocean University of China, Science Press and Springer-Verlag GmbH Germany 2018
s. E-mail: zshen218@126.comE-mail: liuhb@ust.hk
杂志排行
Journal of Ocean University of China的其它文章
- Offshore Fault Geometrics in the Pearl River Estuary, Southeastern China: Evidence from Seismic Reflection Data
- Application of Geoid Anomalies to the Tectonic Research in the East Asian Continental Margin
- Middle Holocene Organic Carbon and Biomarker Records from the South Yellow Sea: Relationship to the East Asian Monsoon
- Mesozoic Deformation and Its Geological Significance in the Southern Margin of the South China Sea
- Optimization of Shanghai Marine Environmental Monitoring Sites in the Identification of Boundaries of Different Water Quality Grades
- Seasonal Variation of Environmental Variables and Phytoplankton Community Structure and Their Relationship in Liaodong Bay of Bohai Sea, China
