Duplex microRNAs assay based on target-triggered universal reporter hybridization
2018-08-22YinanWangYueSunChoiwanLauJianzhongLu
Yinan Wang,Yue Sun,Choiwan Lau,Jianzhong Lu
School of Pharmacy,Fudan University,826 Zhangheng Road,Shanghai 201203,China
Keywords:xMAP array Multiple microRNAs Target-triggered Universal reporter
ABSTRACT In this paper,we designed and evaluated a duplex detection strategy for microRNAs(miRNAs)using universal probe-based target-triggered double hybridization and fluorescent microsphere-based assay system(xMAP array).In the absence of target miRNA,reporter DNA cannot hybridize stably with the immobilized capture DNA due to its low melting temperature.Only after adding target miRNA,can reporter probe hybridize with capture probe to form a stable three-component complex.This targettriggered stable hybridization makes this method possible for highly selective and sensitive detection of multiple miRNAs.We exemplified a quantitative detection of duplex miRNAs with a limit of detection of 40 pM.The xMAP array platform holds the potential of extending this approach to simultaneous detection of up to 100 miRNA targets.Considering the simplicity,rapidity and multiplexing,this work promised a potential detection of multiple miRNA biomarkers for early disease diagnosis and prognosis.
1.Introduction
More than 2500 microRNAs(miRNAs)have been found in humans,and most of them play important regulatory roles in a wide range of biological processes,including cellular differentiation[1],proliferation,apoptosis[2]and development[3].Abundant studies have revealed that miRNAs are associated with various human diseases,including cancers such as breast cancer,ovarian cancer,and lung cancer[4–6],cardiovascular disease[7,8],neurological disease[9],and diabetes[10,11].An ideal diagnostic miRNA assay should be sensitive,specific,simple,rapid,and multiplexed.However,high sensitivity and high specificity of miRNA detection are challenging because of the unique characteristics of miRNA such as the short length of miRNA sequences,the sequence homology among family members,and low abundance[12,13].Current standard miRNA detection strategies include Northern blot,microarray,and reverse-transcription polymerase chain reaction,which generally require large amounts of sample and target amplification,and are labor-intensive,and not easily multiplexed[14,15].
The three-component strategy,which involves a pair of DNA probes(capture and labeled reporter probes)that flank the target sequence,has been widely employed in the DNA and RNA detection assays.However,the short length of miRNAs makes it difficult to form a strong and specific three-component complex.For example,Qiu et al.[16]and Díaz et al.[17]developed Tb-to-QD FRET-based hybridization assays and found that hybridization of both donor and acceptor probes to a single short miRNA strand is not very stable at low concentrations.Arata et al.[18]developed a method for miRNA detection by sandwich hybridization taking advantage of the coaxial stacking effect.They found that the efficiency of hybridization between capture and miRNA is the limiting factor in the sandwich hybridization.To overcome this issue,the direct detection of miRNA was made through the use of fluorescent locked nucleic acid(LNA)probes,whose higher binding affinity to miRNA compared with conventional DNA enabled the use of short probe lengths[19].Sometimes,probe ligation is used to stabilize the miRNA to the capture[20,21].In this regard,our group in 2010 reported a new strategy of template-dependent hybridization for short-length DNA detection based on target-enabled hybridization of a capture probe with a long reporter probe[22].In 2012,we further employed a template-dependent surface hybridization using quantum dotenhanced methodology to detect dual short RNA sequences[23].A multiplexed “mix-and-measure”method would be highly desirable for measuring the expression levels of miRNAs.However,in the template-dependent hybridization assay,any permutation of eightnucleotide tail sequence of the long reporter probe is limited,which causes limit to the simultaneous detection of multiple miRNAs.
The Luminex xMAP array is a microsphere-based multiplex system which can simultaneously detect up to 100 analytes from a single sample.Herein we developed a novel universal probe-based target-triggered double hybridization strategy for the detection of dual miRNAs utilizing xMAP array platform.To achieve this purpose,the corresponding capture probes were immobilized on a specific color-coded microsphere.Each capture probe was designed to have three functional segments,namely 5’poly(A)spacer sequence,middle complementary sequences to specific miRNA,and 3’complementary sequence to a biotin-modified eight nucleotide reporter probe.In the absence of specific target,universal reporter probes and capture probes cannot anneal to each other due to low melting temperature.Only after target miRNA bound to capture recognition section,reporter DNA was allowed to hybridize with capture probe.Thus the biotin labeled on the reporter probe was attached to the microsphere surface,and then reacted with streptavidin-phycoerythrin(SA-PE)to emit fluorescence.Although a duplex analysis is demonstrated here,higher plexing is possible.The proposed method possesses practical adaptability and generality for higher plexing miRNA detection.Only the recognition domain of capture probes needs to be designed according to target sequences,which simplifies the design of higher plexing miRNA detection.The utilization of universal reporter probe in simultaneous assay of multiple miRNAs can also alleviate the cross interference between multiple capture and reporter probes.In view of these advantages,the strategy developed here provides a universal technology for developing simple biosensors for sensitive and selective detection of multiple miRNAs.
2.Materials and methods
2.1.Materials and reagents
All oligonucleotides were obtained from Sangon Biotechnology Co.,Ltd.(Shanghai,China),and miRNAs were synthesized by Takara Biotechnology Co.,Ltd(Dalian,China).The sequences of oligonucleotides used in this work are listed in Table 1.Carboxylated microspheres were purchased from Luminex Corp.(Austin,TX,USA).1-Ethyl-3-(3-dimethylamino-propyl)carbodiimide(EDC)was obtained from Sigma-Aldrich(Shanghai,China).Bovine serum albumin (BSA)wasprovidedby AiziteBiotechnologyCorp.(Shanghai,China).SA-PEwaspurchasedfrom Jackson ImmunoResearch(West Grove,PA).Sheath fluid was procured from Bio-Rad Corp.(Shanghai,China).MiRNeasy mini kit was purchased from QIAGEN(Venlo,Netherlands).All chemicals were of analytical grade and were used as received.All of the solutions were prepared with ultrapure water from a Millipore Milli-XQ system(Bedford,MA,USA),treated with DEPC.
2.2.Preparation of capture DNA-microspheres conjugates
Before conjugation,carboxylated microspheres were activated in 5mg/mL EDC and 0.2M imidazole buffer(pH 6.0)for 20min at 37°C with shaking.Then capture probes were conjugated with activated carboxylated microspheres in 0.2M imidazole buffer for 2h at 37°C with shaking.The reaction volume was proportional to the amount of microsphere.The probe-conjugated microspheres were washed once with wash buffer(7mM Tris-HCl,pH 8.0,0.17M NaCl,and 0.05%Tween 20),and then blocked in buffer A containing 10%BSA for 1h.After that,the supernatant was aspirated and discarded,and the coupled microspheres were resuspended in appropriate volume buffer A(20mM Tris-HCl,pH 8.0,and 0.5M NaCl)to bring the microsphere density at the same level as the stock material before conjugation.The capture probe-conjugated microspheres can be stored at 4°C for a few days before use.For multiplex assay,each type of microspheres was individually conjugated with a specific capture DNA before being pooled together for the remaining of the procedure.In this work,two products of Magplex®microsphere coded 35 and 53 were used.
2.3.Target-triggered universal probe hybridization strategy for the detection of multiple miRNAs
Depending on experiment scale,an appropriate amount of capture probe-conjugated microspheres was removed from storage and transferred to 96-well plates in the amount of 0.5μL stock/well.After magnetic separation,the supernatant was discarded.Mixture of miRNA targets and reporter probes prepared in designated concentration were added into each well(25μL/well,in buffer A),and incubated at the designated temperature for 1h.In a duplex assay,the two microsphere types were pooled at an equal volume before dispensing to the 96-well plate.After that,the microspheres were washed once with 50μL wash buffer.The detection step was performed in 50μL/well of SA-PE prepared in buffer A containing 1%BSA.After 30min of incubation,the microspheres were washed once in 100μL wash buffer,and then re-suspended in 100μL of sheath fluid for analyzing on the Luminex 200 instrument.
2.4.Cell culture and total RNA extract measurement
Human breast cancer cells(MCF-7)were cultured according to the instructions of the American Type Culture Collection.Cells were grown in DMEM (Gibco,penicillin 100U/mL,streptomycin 100μg/mL)plus 10%fetal bovine serum(FBS,Gibco)and maintained at 37°C in a humidified atmosphere of 5%CO2and 95%air.The cells were collected and centrifuged at 3000rpm for 5min in a culture medium,and then washed once with PBS buffer.Total RNA samples were extracted using the QIAGEN miRNeasy mini kit.The extracted RNA was eluted into approximately 50μL of H2O.The total RNA concentration was determined by the UV absorption at 260nm with the following formula:concentration ofRNA sample=40μg/mL×A260×dilution factor.The RNA sample in these cells was diluted to 1μg/μL before analysis.The following procedure of miRNA detection was as described in the target-triggered universal reporter hybridization.

Table 1 DNA and miRNA sequence used in this work.
3.Results and discussion
3.1.Principle of target-triggered universal reporter hybridization
This xMAP array platform based duplex detection strategy for miRNA is demonstrated in Scheme 1.The capture probes consist of three function domains:a poly(A)spacer sequence to extend the capture probe away from the microsphere surface,middle complementary sequences to specific miRNA,and the reporter binding section having the same sequence for the binding of the universal reporter probes.In the absence of target miRNA,the reporter DNA cannot bind with the long capture DNA stably due to short complementary domain(eight base pairs)and low melting temperature,where only an insignificant fluorescence signal was observed at the low intensity of an average~100 fluorescence counts(Fig.1A),showing that the hybridization efficiency of the biotinlabeledreporter probe with immobilizedcapture DNA was extremely low.When target miRNA bound with the target recognition section of capture probes,the reporter DNAs were thus allowed to hybridize stably with complementary capture probe that was immobilized on specific color-coded microspheres.In the presence of 0.4 nM of target miR-21,the signal was increased to an average of~250 fluorescence counts(Fig.1B).When the target miR-21 concentration was increased to 4nM,the signal was increased to an average of~3000 fluorescence counts(Fig.1C).
3.2.The design of the probes
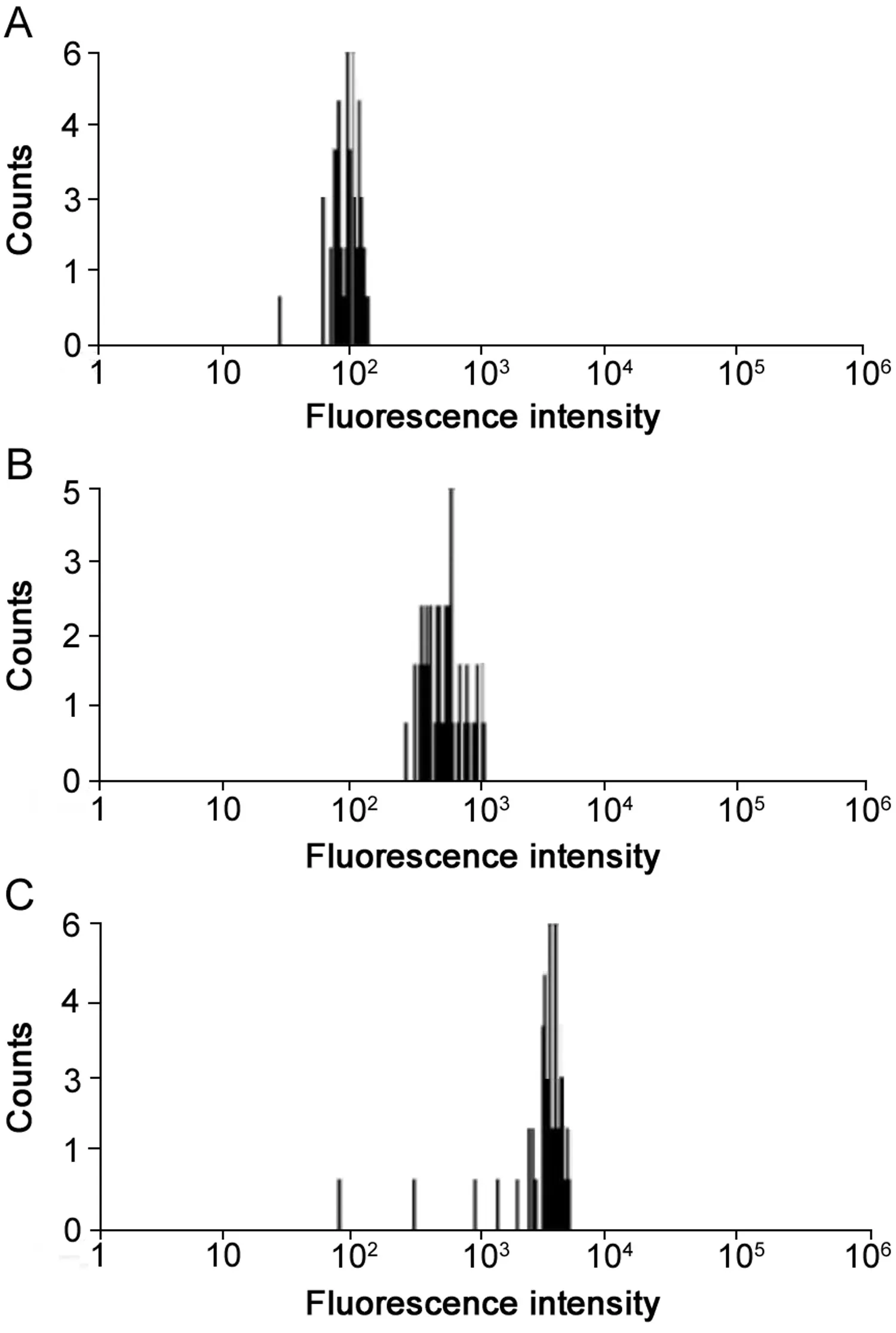
Fig.1.FL intensity comparison.Experimental conditions:20 pmol capture,1 μL MagPlex microsphere stock/well,reporter concentration at 4 nM,miRNA and reporter hybridization temperature at 25 °C,SA-PE:5 μg/mL,miRNA concentration:0 nM(A),0.4 nM(B)and 4 nM(C).
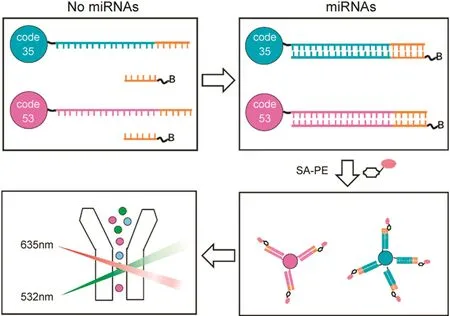
Scheme 1.Schematic illustration of xMAP array based multiplex detection of microRNAs utilizing target-triggered universal reporter hybridization.
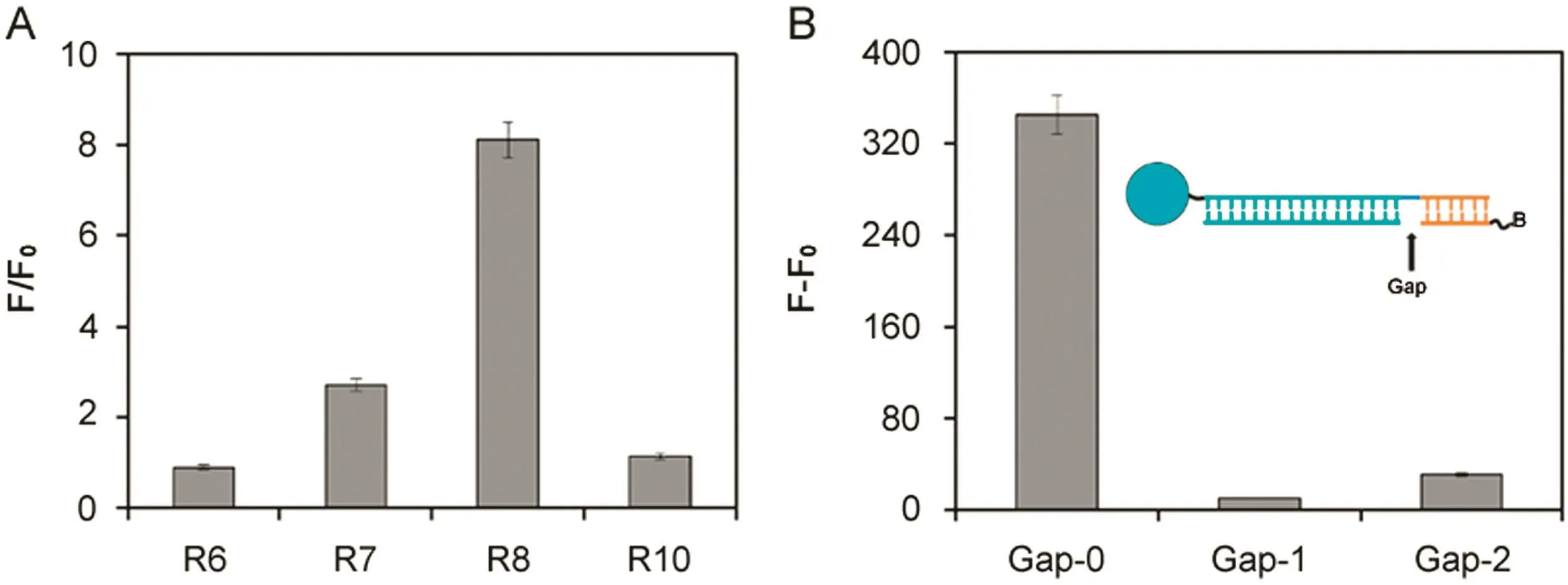
Fig.2.(A)Design of the length of universal reporter probes.Experimental conditions:20 pmol capture-miR-21-gap/well;0.5 μL MagPlex microsphere stock/well;reporter concentration at 20 nM(universal-reporter-6,universal-reporter-7,universal-reporter-8 and universal-reporter-10 were abbreviated to R6,R7,R8 and R10,respectively);miR-21 and reporter hybridization temperature at 25 °C;5 μg/mL SA-PE;and 0.4 nM miR-21.(B)FL intensity vs different gaps between universal reporter probes and targets.Experimental conditions:20 pmol capture-miR-21-gap/well;0.5 μL MagPlex microsphere stock/well;universal-reporter-8 concentration at 20 nM;miR-21 and reporter hybridization temperature at 25 °C;5 μg/mL SA-PE;and 0.4 nM miR-21.
Among factors that affect the DNA duplex stability such as sequence composition,secondary structure,temperature and ionic strength[24],the two most in fluencing factors are the base pairing between complementary strands and stacking between adjacent bases[25].When two contiguous tandem sequences(reporter and target miRNA)are annealed on a longer capture DNA strand,coaxial stacking at the nick brings additional stability and efficiency to the hybridization.The melting temperature of a double strand DNA is proportional to the number of complementary base pairings.Short complementary domain between the capture and reporterDNA results in low hybridization efficiency and insignificant background signal.In our system,“base pairing between complementary strands”is related to the strand length of the reporter-capture probe binding area and the target-capture binding area.Because the length of the target is fixed,the strand length of the target-capture binding area is also pre-determined.Therefore,we first optimized the strand length of hybridization domain between the capture probe and the reporter.The ratio of fluorescence intensity was de fined as F/F0,in which F represents total fluorescence intensity of sample,and F0represents the fluorescence of the blank.The net fluorescence intensity was defined as(F–F0).As shown in Fig.2A,the signal intensity is low when the reporter strand length is too short to stabilize the hybridization complex;while when strand length is too long,the reporter can bind with capture stably even in the absence of target which results in the significant increase of background signal.The optimal length of universal probe was found to be eight nucleotides that produced the best F/F0ratio,and thus was employed in the following study.
We also investigated the gap between the reporter and the target after hybridizing with capture probes.There had been a report proving that continuous stacking hybridization of two duplexes resulted in their mutual stabilization and that the dissociation temperature of the duplex was increased[26].The fluorescence intensity was found to decrease sharply with the introduction of gap nucleotides(Fig.2B)because the coaxial interrupted stacking resulting destabilized three-component hybridization.These gap experiments further confirm that“stacking between adjacent bases”in the presence of target plays a key role in thermodynamic stability of the formed helixes.
3.3.The thermodynamics of the biosensor
Oligonucleotide hybridization is a highly temperature-dependent event.The complementary sequence needs a certain level of thermal energy to bind with each other[27].In the presence of target miRNA,the melting temperature between capture DNA and the reporter DNA is elevated at the beginning,leading to an increase in stability of the complex formed.After the temperature reaches the threshold,the complex hybridization melts and dissociates.Therefore,we investigated both the change of background and target signal when temperature increased from 10 to 35°C(Fig.3A).In the absence of targets,capture probes and reporter probes could form duplex partly at low temperature such as 10°C,resulting in a strong background signal.With the increase of temperature,the destabilization of duplex occurred and the capture and reporter DNA were almost completely separated,and thus an insignificant background signal was seen.After the introduction of target miRNA,initially high fluorescence intensity was observed at low temperatures as a result of the formation of a stable three-component complex.The hybridization efficiency decreased gradually with increasing temperature,which resulted in the decrease of target signal.The optimal F/F0ratio was identified at 25 °C(Fig.3B)for both miRNA assays and thus 25 °C was selected for subsequent experiments.
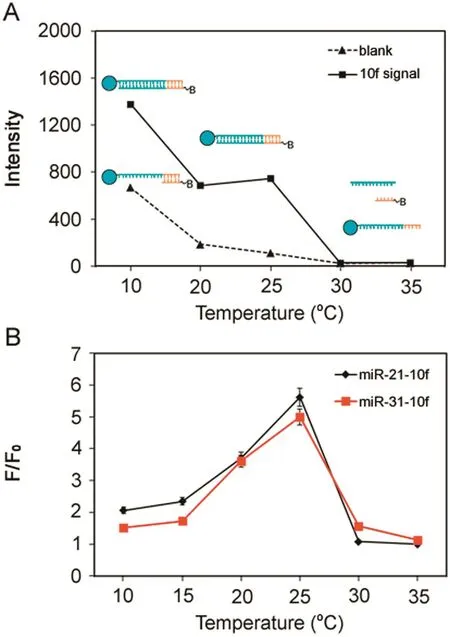
Fig.3.(A)Thermodynamic studies.(B)F/F0ratio vs.the reaction temperature.Experimental conditions,25 pmol capture/well for miR-21,10 pmol capture/well for miR-31,0.5 μL microsphere stock/well,reporter concentration at 20 nM,miRNA and reporter hybridization temperature at 10–35 °C,and 5 μg/mL SA-PE.
3.4.Optimization of reaction parameters
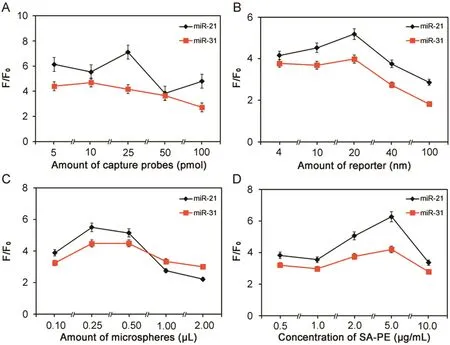
Fig.4.Optimization of capture probe coupling.Experimental conditions:(A)microsphere density was at 0.5 μL stock/well;target concentration at 0.4 nM;reporter concentration at 20 nM;miRNA and reporter hybridization temperature at 25 °C;and 5 μg/mL SA-PE;(B)capture probes:25 pmol for miR-21 and 10 pmol for miR-31;0.5 μL microsphere stock/well;target concentration at 0.4 nM;miRNA and reporter hybridization temperature at 25 °C;and 5 μg/mL SA-PE;(C)capture probes:25 pmol for miR-21 and 10 pmol for miR-31;target concentration at 0.4 nM;reporter concentration at 20 nM;miRNA and reporter hybridization temperature at 25 °C;and 5 μg/mL SA-PE;(D)capture probes:25 pmol for miR-21 and 10 pmol for miR-31;0.5 μL microsphere stock/well;target concentration at 0.4 nM;reporter concentration at 4 nM;miRNA and reporter hybridization temperature at 25°C.
To improve the detection sensitivity,several parameters were investigated systematically to establish optimal conditions for detecting the target miRNAs,including the amount of capture probe and reporter,microsphere density,and SA-PE(Fig.4).In the probe concentration optimization experiment,coupling density ranging from 5 to 100 pmol/well microspheres was investigated.Since the hybridization among target,reporter and capture probes is a dynamic process,the increase of capture density on the surface of coded microspheres promotes both the formation of threecomponent complex and the nonspecific binding of reporter probes.As expected,both the background and target signal increased with the coupling concentration(data not shown).For F/F0,it first increased as capture probes were elevated,and then started to decrease after peaking at 25 pmol for miR-21 and 10 pmol for miR-31,respectively(Fig.4A).The reporter concentration was optimized by screening the reporter concentration from 0.1 to 2.5 pmol.Data from both miR-21 and miR-31 showed that background and target fluorescence intensities increased with the increase of the reporter concentration(data not shown),indicating that reporter probes were a source of non-specific binding in this system.As shown in Fig.4B,the F/F0ratio increased from 4 to 20 nM of reporter in the beginning,and then dropped from 20 to 100 nM of reporter.Based on the F/F0ratio data,20 nM reporter probes were selected for both miR-21 and miR-31.In the detection system,instead of counting the total fluorescence signal of the reaction,the Luminex system software reports the fluorescence intensity on the coded microsphere which represents the medium signal of all counted microspheres.In addition,microspheres density could potentially affect the assay performance,since the fewer microspheres in the reaction,the more target will be captured on a given bead,resulting in higher fluorescent readout.Here,we screened microsphere density in the range of 0.1~ 2μL stock per well(0.1~ 2× [1.25× 104]microspheres/well)and thus 0.25μL stock microspheres per well was selected for subsequent experiments(Fig.4C).At the last step of the assay,SA-PE was the last parameter to be optimized.Various dilutions of SA-PE in buffer A with 1%BSA was tested from 1 to 20μg/mL.The F/F0ratio was increased as the SA-PE concentration increased from 0.5 to 10μg/mL,and then dropped at a higher concentration of SA-PE.Based on the data,5μg/mL SA-PE was selected for future experiments(Fig.4D).
3.5.Interference test
To evaluate the assay specificity,response signal at 0.4 nM or 4 nM of miR-21 and miR-31 was compared in singlex assay(miR-21 or miR-31 only)versus duplex assay(mi-21 and miR-31 together).To ensure side-by-side comparison,both types of microspheres coupled with miR-21 and miR-31 capture DNA sequences were involved in the singlex and duplex assays.The only difference was that only one target miRNA was involved in the singlex assays,while both target miRNAs were present in the duplex assays.Interference was evaluated by comparing the response signal(after background subtraction)in the singlex assay versus that in the duplex assay(Fig.5).For both miR-21 and miR-31,comparable signal was observed in the singlex and duplex assays,proving that no interference occurred when the two targets were measured simultaneously.
3.6.Analytical performance
Under the optimized conditions,the sensitivity and dynamic range of the multiplex assays was examined with different target miRNA concentrations.As shown in Fig.6,a good correlation between fluorescence intensity and miRNA concentration was observed from 40 pM to 4nM for both target miRNAs.A low limit of detection at 40 pM was achieved for both target miRNA.The applicability of the developed approach for miRNAs analysis in real samples was tested by monitoring the two target miRNAs in total RNA samples extracted from cancer cells.The endogenous levels of miR-21 were detected to be 2.64 ± 0.71 fmol/μg/μL total RNA extract in breast cancer MCF-7 while the miR-31 was not found in either cancer cells.The sample analysis was also validated by a reference method,i.e.poly(A)tailed real-time PCR method.Reverse-transcription and qRT-PCR procedure were performed by all-in-one miRNA qRT-PCR detection system from GeneCopeia(Guangzhou,China)according to the manufacturer's instructions.Comparableresultswereobtained with standard qRT-PCR(2.45 ± 0.12 fmol for miR-21 in 1μg/μL total RNA extract from MCF-7).It can be concluded that the presented technique proves to be a potential detection of multiple miRNA biomarkers for early disease diagnosis and biomedical research.
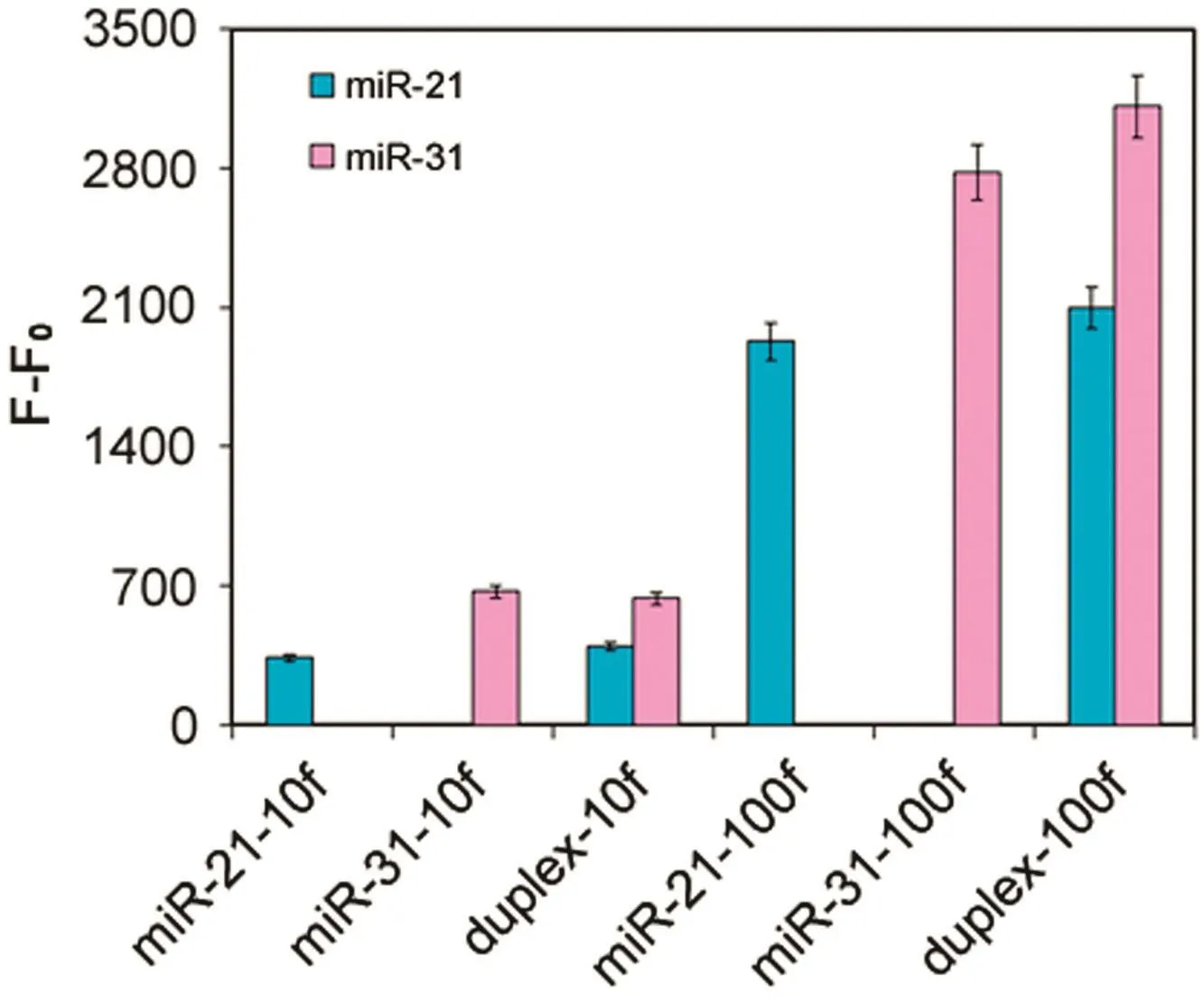
Fig.5.Interference tests.Experimental conditions:capture probes 25 pmol for miR-21;10 pmol for miR-31;0.5 μL microsphere stock/well;target concentration at 0.4 nM or 4 nM;reporter concentration at 20 nM;miRNA and reporter hybridization temperature at 25 °C;and 5 μg/mL SA-PE.
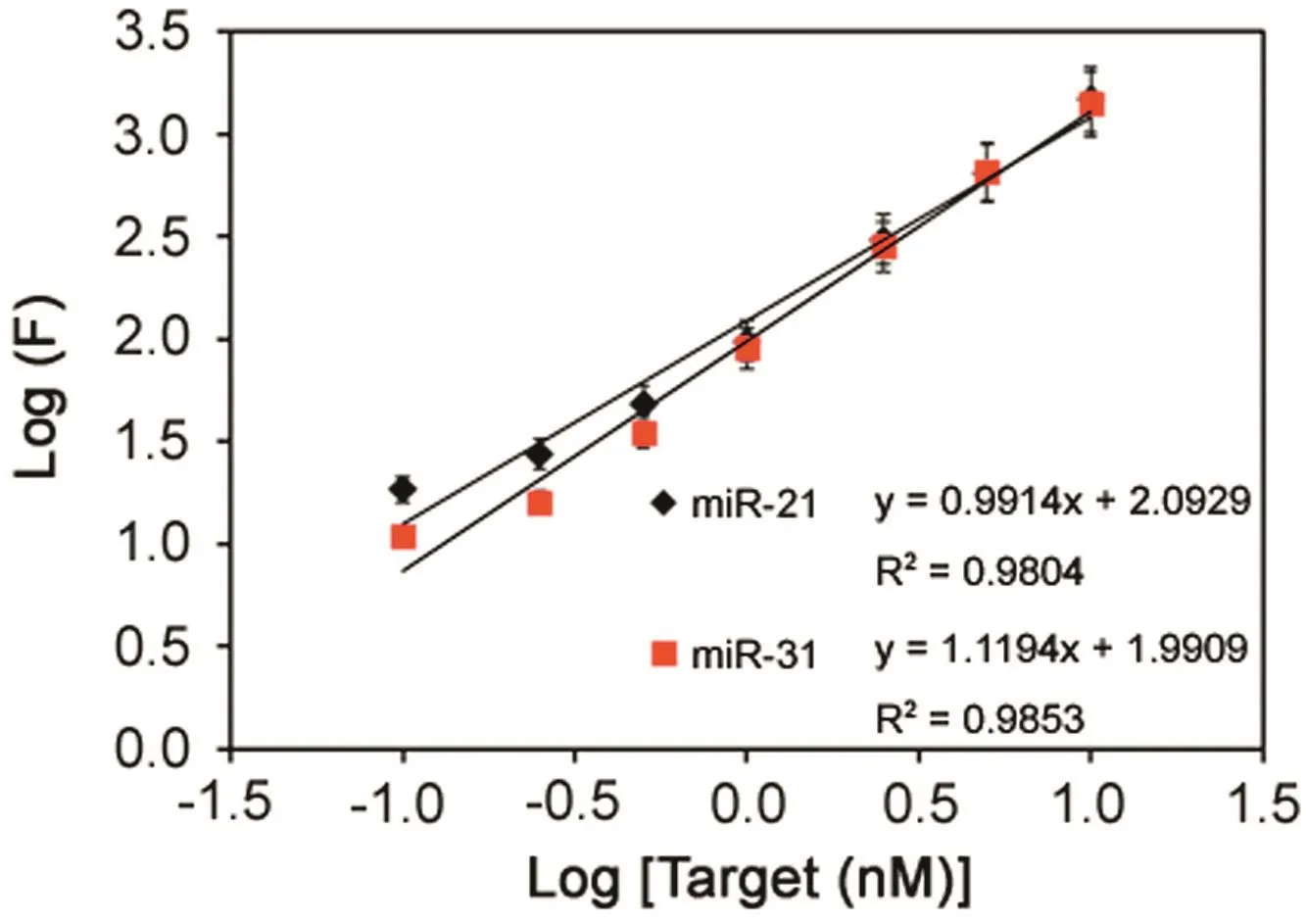
Fig.6.Log-log calibration data for simultaneous detection of miR-21 and miR-31.Experimental conditions:0.25 μL microsphere stock/well;reporter concentration at 20 nM;miRNA and reporter hybridization temperature at 25 °C;and 5 μg/mL SA-PE.
4.Conclusion
A simple and sensitive method for detection of multiple miRNAs based on xMAP array and target-triggered universal reporter hybridization was established.The method required the recognition of the target to promote the reporter efficient annealing with magnetic bead-attached capture DNA.The strategy was successfully exemplified for duplex detection of microRNAs in a single sample without any additional amplification processes.The targettriggered universal reporter hybridization assay can be easily applied in higher plexing with less cross interference and no need for complicated probe designation.In view of these advantages,the proposed assay could create a universal technology for developing simple biosensors for sensitive and selective multiplex detection of short-length DNA and small RNA,which is promising in miRNA expression profiles and clinical diagnosis.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was financially supported by the National Science Foundation of China(Grant No.21575029).
杂志排行
Journal of Pharmaceutical Analysis的其它文章
- Advances in tumor-endothelial cells co-culture and interaction on micro fluidics
- Analytical methods for investigating in vivo fate of nanoliposomes:A review
- Recent advances in screening of enzymes inhibitors based on capillary electrophoresis
- Unusual retention behavior of omeprazole and its chiral impurities B and E on the amylose tris(3-chloro-5-methylphenylcarbamate)chiral stationary phase in polar organic mode
- Screening potential mitochondria-targeting compounds from traditional Chinese medicines using a mitochondria-based centrifugal ultra filtration/liquid chromatography/mass spectrometry method
- Simultaneous determination of steroid drugs in the ointment via magnetic solid phase extraction followed by HPLC-UV
