Molecular cloning and transcriptional analysis of a NPY receptor-like in common Chinese cuttlefishSepiella japonica*
2018-07-11YANGJingwen杨静文XUYuchao徐玉超XUKe许珂PINGHongling平洪领SHIHuilai史会来Zhenming吕振明WUChangwen吴常文WANGTianming王天明
YANG Jingwen (杨静文) XU Yuchao (徐玉超) XU Ke (许珂)PING Hongling (平洪领) SHI Huilai (史会来) LÜ Zhenming (吕振明)WU Changwen (吴常文) WANG Tianming (王天明)
1National Engineering Research Center for Facilitated Marine Aquaculture,Marine Science College,Zhejiang Ocean University,Zhoushan 316022,China
2Marine Fisheries Research Institute of Zhejiang Province,Zhoushan 316022,China
AbstractNeuropeptide Y (NPY) has a pivotal role in the regulation of many physiological processes.In this study, the gene encoding a NPY receptor-like from the common Chinese cuttlefishSepiella japonica(SjNPYR-like) was identified and characterized. The full-lengthSjNPYR-likecDNA was cloned containing a 492-bp of 5′ untranslated region (UTR), 1 182 bp open reading frame (ORF) encoding a protein of 393 amino acid residues, and 228 bp of 3′ UTR. The putative protein was predicted to have a molecular weight of 45.54 kDa and an isoelectric point (pI) of 8.13. By informatic analyses,SjNPYR-like was identified as belonging to the class A G protein coupled receptor (GPCR) family (the rhodopsin-type). The amino acid sequence contained 12 potential phosphorylation sites and five predicted N-linked glycosylation sites.Multiple sequence alignment and 3D structure modeling were conducted to clarifySjNPYR bioinformatics characteristics. Phylogenetic analysis identifi es it as an NPYR with identity of 33% toLymnaea stagnalisNPFR. Transmembrane properties ofSjNPYR-like were demonstrated in vitro using HEK293 cells and the pEGFP-N1 plasmid. Relative quantifi cation ofSjNPYR-like mRNA level confi rmed a high level expression and broad distribution ofSjNPYR-likein various tissues of femaleS.japonica. In addition, the transcriptional profi le ofSjNPYR-likein the brain, liver, and ovary during gonadal development was analyzed. The results provide basic understanding on the molecular characteristics ofSjNPYR-like and its potentially physical functions.
Keyword: Sepiella japonica; NPY receptor-like; growth; reproduction; gene expression
1 INTRODUCTION
Neuropeptides are small protein-like molecules used by neurons to transmit important regulatory signals in both vertebrates and invertebrates. Several neuropeptides are widely conserved in the animal kingdom from protostomes to deuterostomes(Blumenthal, 2010; Minakata, 2010; van Loy et al.,2010; Nässel and Wegener, 2011; Grimmelikhuijzen and Hauser, 2012). The neuropeptide Y (NPY)superfamily, including pancreatic polypeptide (PP)and peptide YY (PYY), is responsible for the central regulation of multiple physiological processes in vertebrates, such as food intake, energy balance,learning, and others (Redrobe et al., 1999; Beck,2001; Hökfelt et al., 2008; Nguyen et al., 2011). The identifi cation ofits invertebrate counterpart showed that these NPY homologs have a characteristic C-terminus ending with an amidated Phe (F) rather than a Tyr (Y) residue and, hence are designated Neuropeptide F (NPF). The regulatory roles of NPFs in feeding, energy homeostasis, reproduction, and stress responses have been demonstrated in a variety ofinvertebrates including mollusks (Suzuki et al.,2002; Nässel and Wegener, 2011). Recently, NPF has been identified in numerous molluscan species such as the garden snailHelixaspersa(Leung et al., 1992),the marine molluskAplysiacalifornica(Rajpara et al., 1992), the squidLoligovulgaris(Smart et al.,1992), the gastropod snailLottiagigantea(Veenstra,2010), the pond snailLymnaeastagnalis(de Jong-Brink et al., 2001), and so on. The signaling pathway activated by this neuropeptide is now becoming a new subject for studies to explore the functional regulatory mechanism.
The peptides of the NPY family display a wide array of biological activities that are thought to be mediated through a subfamily of G protein-coupled receptors (GPCRs) termed NPY receptors (Gerald et al., 1996). Five NPY receptors (NPYRs) have been identified from mammals to date: Y1, Y2, Y4, Y5, and Y6. In addition another receptor subtype (Y3) was suggested by pharmacological studies without supporting data from molecular cloning (Michel et al., 1998). Seven totally different NPY receptors have been described in vertebrates (Sundström et al.,2013). The evolution of NPYRs shows that vertebrate ancestors probably had three receptor subfamilies.The subfamily of Y1 includes the Y1, Y4, Y6, and Y8 receptors, the subfamily of Y2 is comprised of Y2 and Y7 (identified in zebrafish and frogs), and the subfamily of Y5 consists of only Y5, due to lack of close relatives of this receptor (Larhammar and Salaneck, 2004). In invertebrates, the NPYRs (also named NPFRs for their ligand NPFs) have been identified functionally for the insectsDrosophila melanogaster,Bombyxmori, andAnophelesgambiae(Garczynski et al., 2002, 2005; Deng et al., 2014) and the mollusksL.stagnalis(Tensen et al., 1998) with additional predicted NPYRs from genomic sequences ofCrassostreagigas(Zhang et al., 2012) andOctopus bimaculoides(Albertin et al., 2015). With limited exploration of the NPYRs, the mechanisms by which NPY/NPYRs regulate multiple physiological activities in mollusks have not been discovered.
The cuttlefishSepiellajaponica(Phylum:Mollusca, Class: Cephalopoda) is distributed mainly in coastal regions of Zhejiang and Fujian provinces,and is now becoming an important aquatic species.Recently, several studies have focused on the reproduction and developmental regulation ofS.japonica(Furuya, 2008; Cao et al., 2016; Lü et al.,2016a, b; Yan et al., 2016). Here, we describe the cDNA cloning of aS.japonicaNPYR-like (SjNPYR-like) that is closely related in sequence toL.stagnalisNPFR and the vertebrate NPY receptor type 2. Further bioinformatic analysis was conducted, and cellular location in vitro and transcriptional detection ofSjNPYR-likein different tissues and developmental stages were investigated. The results suggest thatSjNPYR-like is phylogenetically close to the Y2 family, has the typical characteristics of a transmembrane GPCR, and mediates physiological functions in feeding and energy homeostasis during development.
2 MATERIAL AND METHOD
2.1 Sample collection
Common Chinese cuttlefishS.japonicawas collected from the culture pool at aquaculture station of the Marine Fisheries Research Institute of Zhejiang on the island of Xishan, Zhoushan, Zhejiang Province,China. To evaluate the expression profi le ofSjNPYR-likein different tissues and at different stages during the female reproductive cycle, ovaries were collected at four gonadal development stages categorized using a histological method. The different gonadal development stages (period I—oogonium production period, period III—interstitial growth period, period II—protoplasmic growth period, and period IV—trophoplasmic growth period) are defi ned following the published article (Yan et al., 2016). Different tissues such as brain, liver, gill, branchial heart, and muscle of females in period II, as well as brain, liver,and ovary of females in all four stages (periods I to IV) were collected for further gene expression analysis. All collected tissues were immediately frozen and stored in liquid nitrogen until RNA extraction was performed.
2.2 RNA extraction and cDNA synthesis using rapid amplifi cation of cDNA ends (RACE)
The total RNA for the 5′ RACE assay was extracted from the liver ofS.japonica. RNA was extracted using Trizol reagent (TaKaRa, Japan) and phenol/chloroform. 10.0 mg total RNA were used to perform the 5′ RACE protocol and 1.0 mg was used for the 3′RACE protocol using FirstChoice®RLM-RACE Kit(Life Technologies, Madison, WI, USA) following the manufacturer’s instructions. The cDNA products for the next PCR steps were stored at -20°C. The Specific primers used for the RACE assays are listed in Table 1.

Table 1 Sequences of primers used for 5′ and 3′ RACE of S. japonica NPYR gene and gene expression analysis
2.3 Sequence analysis
Comparison of the cDNA sequences with known sequences published in GenBank to determine percent identity was computered by online BLAST tools(BLASTX 2.5.1+; http://blast.ncbi.nlm.nih.gov). The encoding sequence of theSjNPYR-like was translated into amino acid sequence by BioEdit 7.0.5.3. The N-glycosylation sites were predicted using the NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/) and the phosphorylation sites were deduced by NetPhos 2.0 Server (http://www.cbs.dtu.dk/services/NetPhos/). The physicochemical properties of these proteins were predicted using ProtParam (http://www.expasy.org/tools/protparam.html). The predicted amino acid sequence of theSjNPYR-like and other homologs were aligned using online Pairwise Sequence Alignment tool (http://www.ebi.ac.uk/Tools/psa/). The transmembrane region ofSjNPYR-like was predicted using TMpred(http://www.ch.embnet.org/software/TMPRED_form.html). The prediction of protein domains were performed using online SMART (http://smart.emblheidelberg.de/) and InterProScan (http://www.ebi.ac.uk/InterProScan/). The NPYR 3D structure was modeled using SWISS-MODEL (http://swissmodel.expasy.org/). Analysis of the protein secondary structure was done using PredictProtein (http://www.predictprotein.org/). Based on the Maximum Likelihood (ML) Statistical Method, the phylogenetic tree was constructed using MEGA 5.1.
2.4 Mammalian expression vectors construction
To amplify the full-length encoding sequence ofSjNPYR-like, primers were designed based on its full-length cDNA sequence and to allow for subcloning into the pEGFP-N1 plasmid (Table 1).The PCR products were inserted into the fi nal pEGFP-N1 expression vector using the HidIII and KpnI restriction enzymes (Beyotime, Haimen, China)and Rapid DNA Ligation Kit (Beyotime, Haimen,China). The constructed vector was namedSjNPYR-like-EGFP, and sequenced to verify the correct reading frame.
2.5 Transfection and confocal microscopy
The HEK293 cells (human embryonic kidney cell line) was cultured in equilibrated growth medium(Dulbecco’s modified Eagle’s medium (DMEM),10% fetal bovine serum (FBS, HyClone, Logan, UT,USA), 4 mmol/L-glutamine (Invitrogen, Madison,WI, USA)) at 37°C in a humidified atmosphere containing 5% CO2.SjNPYR-like-EGFP was transfected into HEK293 cells using with X-tremeGENE HP DNA transfection reagent (Roche Applied Science, Indianapolis, USA) following to the manufacturer’s instructions. After 12-16 h, the transfected HEK293 cells were seeded onto glass coverslips. After 16-24 h, cells were stained using DiI as the cell membrane probe (Beyotime, Haimen,China) for 5-10 min, then fi xed with 4%paraformaldehyde at room temperature for 15 min,and fi nally stained by DAPI (Beyotime, Haimen,China) for 10 min incubating.
2.6 Real-time quantitative PCR (qRT-PCR)
Total RNA from brain, liver, ovary, gill, branchial heart, and muscle tissues was extracted from female cuttlefishes in late vitellogenic stage. Moreover, brain,liver and ovary were collected at four gonadal development stages. Reverse transcription was conducted using the M-MLV reverse transcriptase(TaKaRa, Japan) and products was kept at -20°C for the qRT-PCR analysis.β-actinwas chosen as the internal control (housekeeping) gene. Specific primer sequences forβ-actinwere from previous reports (He et al., 2014; Yan et al., 2016). qRT-PCR primers Specific forSjNPYR-like(Table 1) were designed according to theSjNPYR-likeCDS sequence. The qRT-PCR assays were carried out by the ABI 7500 Software v2.0.6 (Applied Biosystems) and performed using the SYBR PrimeScript™ RT reagent Kit(TaKaRa, Japan).
The relative transcriptional level ofSjNPYR-likewas calculated using the 2-ΔΔCtmethod (Livak and Schmittgen, 2001). All data are presented as mean±S.D. (standard deviation). Differences were tested using PASW Statistics 18.00 (SPSS Inc.,Chicago, IL, USA) with one-way Analysis of Variance(ANOVA) followed by Tukey’s post hoc test. The signifi cance level was set at 0.05.
3 RESULT
3.1 Isolation and characterization of SjNPYR- like cDNA
The full-length cDNA sequence ofSjNPYR-like,1 902 bp long, was cloned fromS.japonica(GenBank accession No. KX683395). The sequence contained an open reading frame (ORF) of 1 182 bp encoding a protein of 393 amino acids, with 492 bp of 5′untranslated region (5′ UTR), and 228 bp of 3′ UTR(Fig.1). The predicted protein had a theoretical molecular weight of 45.54 kDa and an isoelectric point (pI) of 8.13. The putativeSjNPYR-like was classified as belonging to the class A GPCR family according to the predicted 7TM_1 domain in amino acid sequence of this receptor. Meanwhile, theSjNPYR-like amino acid sequence contained several potential sites for modifi cation (Fig.1). Five potential N-linked glycosylation sites were present, with three in the extracellular N-terminal domain (at amino acids 2, 10, and 14) and two in the intracellular C-terminal domain (aa 339 and 365). Two conserved cysteine residues were present at positions 105 and 182 within the EC1 and EC2 domains. Twelve potential phosphorylation sites were present at five Serine residues (aa 115, 231, 234, 236, and 349), two Threonine residues (aa 96 and 379), and five Tyrosine residues (aa 15, 180, 309, 343, and 374).
The deduced amino acid sequence ofS.japonicaNPYR-like was compared with those of nine other homologous NPYR sequences (NPFRs, NPY2Rs,and NPY7Rs) that varied in length from 336 to 481 amino acid residues. Pairwise ClustalW analysis of amino acid sequences was carried out to evaluate homology relationships. The predictedS.japonicaNPYR-like amino acid sequence showed high identity to the predicted NPY2R sequence ofOctopus bimaculoides(81%), and other functionally identified NPFR and NPYR amino acid sequences, albeit with lower identity (ranging from 29% to 39%). Multiple sequence alignment analysis revealed conservation in the seven transmembrane (7TM) domains of NPYR sequences from various species includingS.japonica(Fig.2). Apart fromS.japonicaNPYR andO.bimaculoidesNPY2R, significant variation was seen in the lengths of the N terminal, EC2, IC3, and C terminal regions (Fig.2). The protein structure ofSjNPYR-like was predicted using SWISS-MODEL(Fig.3a) while secondary structure was predicted by PredictProtein (Fig.3b). Homology modeling revealed that this protein was similar to human OX2 orexin receptor (4s0v.1.A) in the Protein Data Bank. The protein binding regions were predicted and marked on the constructed model.
To examine the relationship ofSjNPYR-like with NPYRs from various other species, neuropeptide FF receptors (NPFFRs) and Pyroglutamylated RFamide peptide receptors (QRFPRs), a phylogenetic tree was constructed with Mega 5.1 using the ClustalW multiple alignment and the protein sequences ofSjNPYR-like and 45 alternate NPYRs, 6 NPFFRs and 6 QRFPRs from the gene bank (Fig.4). Vertebrate NPYRs are categorized into three major clusters corresponding to the Y1 subfamily (NPY1R, NPY4R,and NPY6R), the Y5 subfamily (NPY5R), and the Y2 subfamily (NPY2R and NPY7R). Invertebrate NPYRs were separated into two distinct groups and were relatively close to the vertebrate Y2 subfamily.NPFFRs and QRFPRs clustered together into a seperated group. The deducedSjNPYR-like protein sequence grouped with theO.bimaculoidesNPY2R and formed a cluster with the functionally identified NPFRs fromD.melanogaster,B.mori, andA.gambiae. Meanwhile, theL.stagnlisNPFR and predicatedC.gigasNPYR formed the second group ofinvertebrate NPYRs.
3.2 Expression of Sj NPYR-like-EGFP in HEK293 cells

Fig.1 Full-length cDNA and deduced amino acid sequences of Sj NPYR-likeThe predicted 7TM_1 conserved domain is shaded in grey. The conserved transmembrane domain regions (TM1-TM7) are noted with black underline. The putative N-glycosylated sites and phosphorylation sites are marked with black triangles and boxed by full lines, separatively. The termination codon is marked with an asterisk.
To confi rm the cellular membrane location ofSjNPYR-like, the enhanced green fluorescent protein(EGFP) was fused to the C terminus of the receptor,and the resulting construct was expressed in human embryonic kidney 293 (HEK293) cells. significant expression of this recpetor on cell surface was observed under confocal microscopy (Fig.5). Result suggests the location ofSjNPYR-like in the cell membrane in HEK293 as a transmembrane receptor,and the C-terminal EGFP tag does not affectSjNPYR-like expression.

Fig.2 Alignment of the deduced Sj NPYR-like amino acid sequence with sequences of other speciesSequences ofOctopus bimaculoidesNPY2R (ObNPY2R),Lymnaea stagnalisNPFR (LsNPFR),Drosophila melanogasterNPFR (DmNPFR),Anopheles gambiae(AgNPFR),Bombyx moriNPFR (BmNPFR),Homo sapiensNPY2R (HsNPY2R),Epinephelus coioidesNPY2R (EcNPY2R),Takifugu rubripesNPY7R (TrNPY7R), andCallorhinchus miliiNPY7R (CmNPY7R) were obtained from GenBank database (Suppl. Table 1). The mutiple sequences alignment was prformed using online Pairwise ClustalW. The conserved transmembrane domain regions (TM1-TM7) are noted with black lines above. The extracellular and intracellular domains (EC, IC) are marked above. The black triangles indicate predicted phosphorylation sites and black spots indicate predicted N-linked glycosylation sites.

Fig.3 Predicted Sj NPYR-like protein structure and domain organizationa. predicted 3D structure of theSjNPYR-like protein. The conserved transmembrane domain regions (TM1-TM7), three extracellular and intracellular domains (EC, IC) are noted. The putative 3D structure of theSjNPYR-like protein was modelled by online SWISS-MODEL server; b. the conserved transmembrane regions and protein binding regions ofSjNPYR-like. The dashboard overview was generated by online PredictProtein server.
3.3 Quantifi cation of SjNPYR- like expression at different developmental stages
Different tissues ofS.japonicawere sampled at four stages during the gonadal development process,and body mass and ovarian wet weight were recorded(Fig.6). From relative quantifi cation analysis (Fig.7),SjNPYR-likeexpression was ubiquitous in multiple tissues during the interstitial growth period (III).SjNPYR-likeexpression was high in the brain, liver,and ovary, but low in the gill, branchial heart, and muscle. To determine the expression profi le ofSjNPYR-likein the brain, liver, and ovary tissues ofS.japonicaduring different gonadal development stages, theSjNPYR-likemRNA level was determined(Fig.8). In all three tissues, the transcriptional level ofSjNPYR-likewas relatively high during the protoplasmic growth period (II) and the interstitial growth period (III), and then decreased to low levels in the trophoplasmic growth period (IV), with an extremely low gene expression level in the ovary during the trophoplasmic growth period (IV).

Fig.4 Phylogenetic tree of Sj NPYR-like amino acid sequences with 45 related NPYR homologs, 6 NPFFRs and 6 QRFPRsThe tree was generated by MEGA 5.1 software based on Maximum Likelihood (ML) algorithms with 1 000 bootstrap replications. Y1R: neuropeptide Y1 receptor; Y2R: Y2 receptor; Y4R: Y4 receptor; Y5R: Y5 receptor; Y6R: Y6 receptor; Y7R: Y7 receptor; Y1: Y1 subfamily; Y2: Y2 subfamily; Y5:Y5 subfamily; NPFR: neuropeptide F receptor; NPFFR: neuropeptide FF receptor; QRFPR: Pyroglutamylated RFamide peptide receptor. * indicates the invertebrate NPYRs (NPFRs) which have been identified by functional criteria. Accession numbers are listed in Suppl. Table 2.
4 DISCUSSION
The common cuttlefishS.japonica, distributed mainly in the coastal regions of Zhejiang and Fujian provinces, is an important aquatic species harvested in China (Wu and Tang, 1990; Wu et al., 2010). To improve production, artifi cial breeding methods have been developed, and these aquaculture techniques have been applied successfully in China. However, precocious puberty of culturedS.japonicabecame apparent, and weakened the development of this industry along the coast of China. A good understanding of the regulation of growth and reproduction in this species has become increasingly important. Recent studies on the control of growth and reproduction in cephalopods suggest that NPYR and its ligand NPY play important roles in feeding behavior and regulation of sexual maturation via central neuroendocrine control (Di Cristo, 2013).However, there are no publications on the regulatory roles of the NPY/NPYR system inS.japonica.
This study reports for the fi rst time cloning of the NPYR homolog ofS.japonica. The NPYR cDNA sequence contained 393 amino acid residues (Fig.1).The predicted conserved protein domain 7TM_1 of this putative amino acid sequence indicatedSjNPYR-like belongs to the class A (rhodopsin) GPCR family(Marchler-Bauer et al., 2015). In addition, five N-linked glycosylation sites and 12 phosphorylation sites were predicted. These putative sites are suggested mainly related with the protein activity, function,diversity, localization, cell signaling, and transcriptional modifi cation (Arnold et al., 2007;Miedlich and Abou-Samra, 2008). Further multiple sequence alignment analysis indicated thatSjNPYR-like was conserved with other invertebrate NPYRs and vertebrate Y2 receptors, especially in the transmembrane regions (Fig.2). Otherwise, lower identity in the C-terminal and N-terminal regions of NPYRs from different species could be related to the diverse ligands and signal transduction properties associated with the different receptors. The high conservation betweenSjNPYR-like and putativeObNPY2R of the entire protein sequences, N-linked glycosylation sites, and phosphorylation sites suggested these two receptors belong to the same subtype.
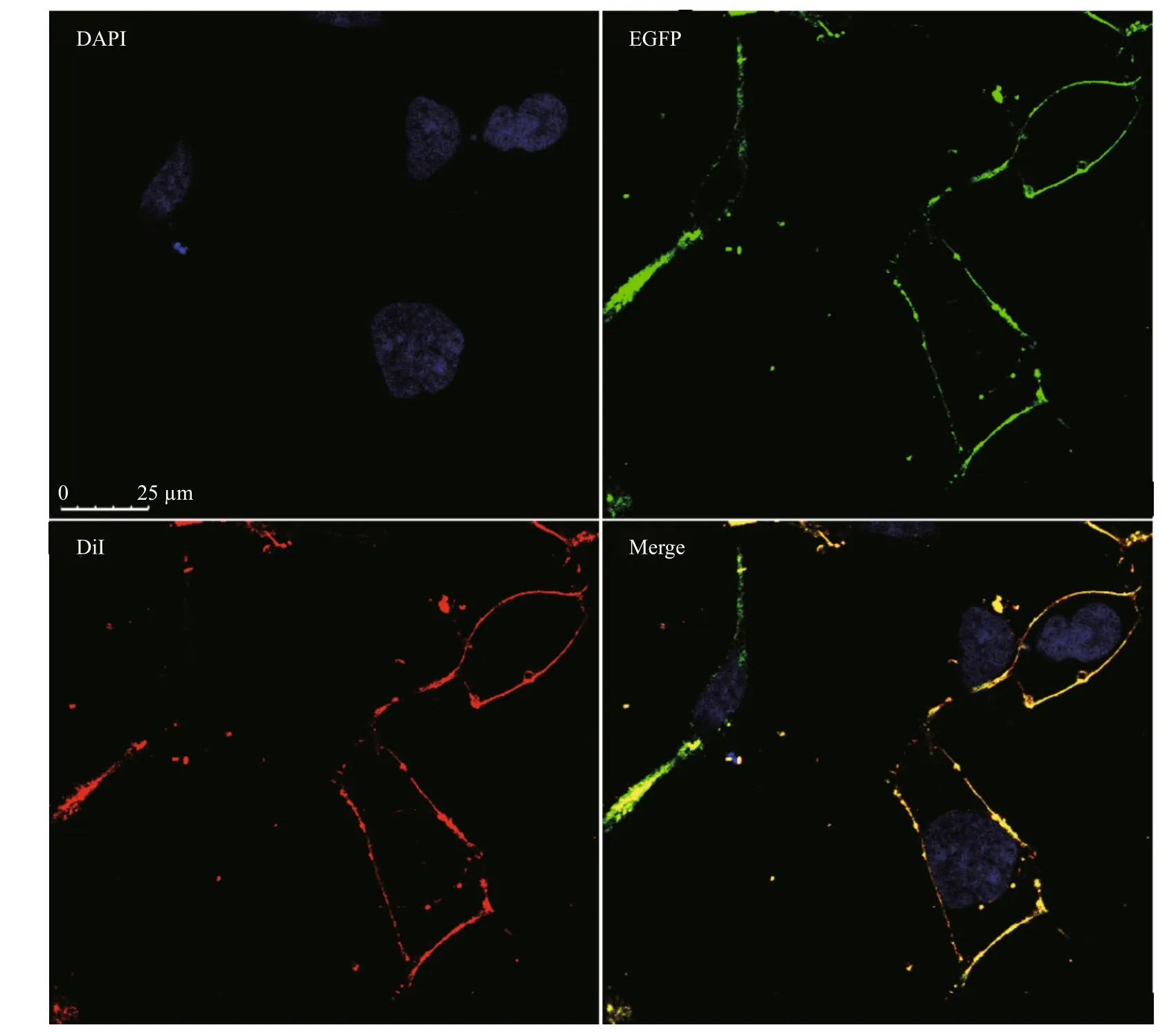
Fig.5 Confocal microscopy of HEK293 cells expressing the Sj NPYR-like-EGFP fusion proteinTransiently expressingSjNPYR-like-EGFP cells were stained with cell membrane probe (DiI) and cell nucleus probe (DAPI), and detected by confocal microscopy. All images are representative of at least three independent experiments.

Fig.6 The body mass and ovarian wet weight during gonadal developmentTissues were collected from females in different gonadal development stages: (I) oogonium production period, (II)protoplasmic growth period, (III) interstitial growth period, and(IV) trophoplasmic growth period. Values indicate mean±S.D.n=6.
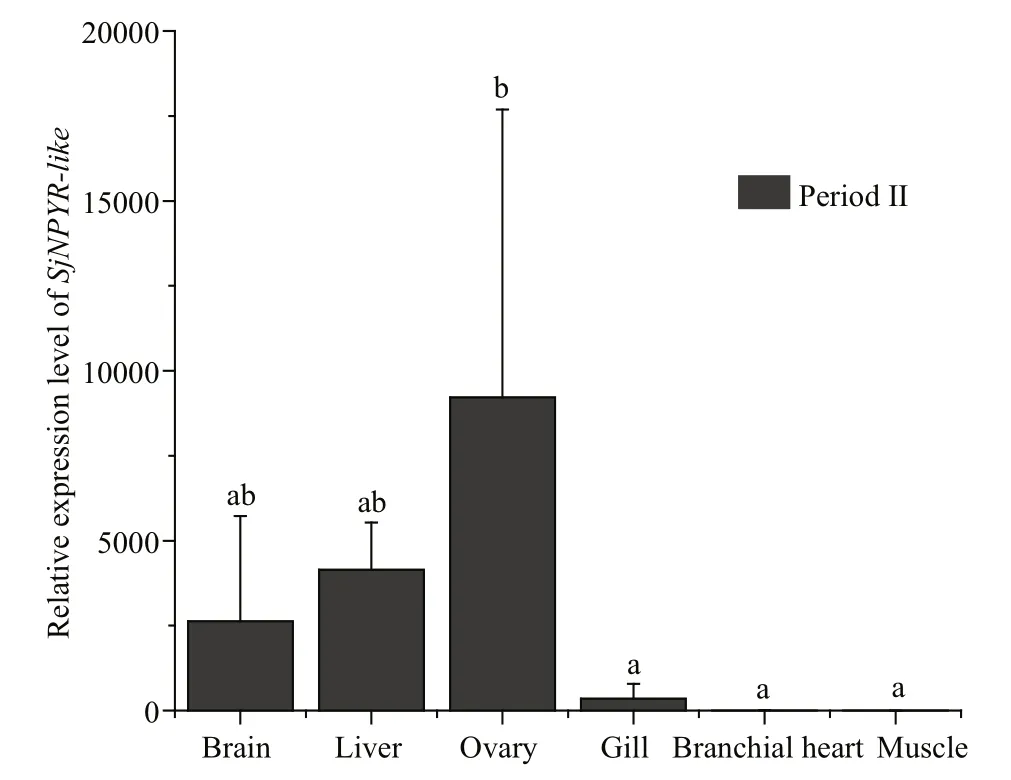
Fig.7 Relative expression of SjNPYR- like in different tissues of females in period II (protoplasmic growth period)Total RNA was isolated and purified from brain, liver, ovary, gill,branchial heart, and muscle. The relative expression value was normalized against the expression ofβ-actin(the housekeeping gene). Different lowercase letters noted above the value bars indicate the significant differences between different tissues (P<0.05).
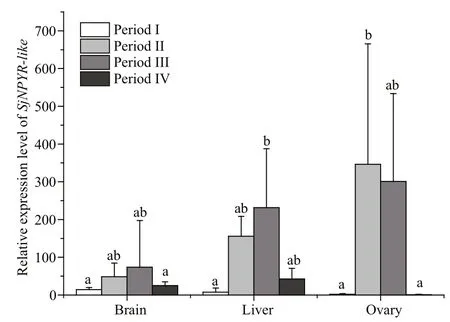
Fig.8 Relative expression of SjNPYR- like in brain, liver,and ovary tissues from females in different gonadaldevelopment stagesI: oogonium production period; II: protoplasmic growth period; III:interstitial growth period; IV: trophoplasmic growth period. The relative expression value was normalized against the expression ofβ-actin(the housekeeping gene). Different lowercase letters above the value bars indicate the significant differences between different tissues (P<0.05).
NPYRs mediate cell signal transduction through binding to the agonist NPYs and activating the coupled G protein. significant progress in research on GPCRs, the ligand-binding modes, receptor activation, and regulation of subsequent internalization mechanisms has been made by many organizations.The intracellular loops and C-terminus are involved in G protein-mediated signaling, internalization, and phosphorylation (Duvernay et al., 2004; Kristiansen,2004). The extracellular loops and N-terminus are probably necessary for ligand recognition and binding(Merten et al., 2007). In this study, bioinformatics analysis was used to model theSjNPYR-like 3D structure and predict its secondary structure. Since the NPYR crystal structure has not been determined,homology of the predictedSjNPYR-like model with the 4s0v.1.A structure (human OX2 orexin receptor)from the Protein Data Bank was analyzed (Fig.3a).SjNPYR-like protein binding regions were analyzed,and potential sites that may be involved with ligand binding were predicted: two (aa 175-177 and 187-190) in the EC2 domain, and one (aa 282-283) in the EC3 domain. Furthermore, regions that are thought to play key roles in G protein coupling were predicted(Fig.4b): four sites (aa 223, 234-236, 239-240, and 245) in the IC2 domain, and one site (aa 329) in the intracellular C-terminal tail.
Phylogenetic analysis was carried out to investigate the evolutionary relationships betweenSjNPYR-like and NPYRs from vertebrates and invertebrates, as well as 6 NPFFR sequences and 6 QRFPR seuqences were also included in this phylogenetic tree to see if the cuttlefish sequences clearly belongs to the NPYR clade to the exclusion of other receptor families(Fig.4). Vertebrate NPYRs clustered into three major groups corresponding to the Y1, Y2, and Y5 subfamilies. Invertebrate NPYRs were separated into two distinct groups and were relatively close to the vertebrate Y2 subfamily. The NPFFRs and QRFPRs clustered together with two seperated groups.However, further studies on theL.stagnalisNPYR(NPFR) suggested that this invertebrate NPYR resembles the Y1 subtype more than the Y2 subtype(Tensen et al., 1998). In addition, theDrosophila melanogasterNPYR (NPFR) was suggested to be not closely related to the Y2 subtype (Garczynski et al.,2002). This indicates experimental data on the signaling pathway characteristics ofinvertebrate NPYRs is crucial for subtype classifi cation. TheSjNPYR-like clustered with the putative NPY2R ofO.bimaculoideswith a high sequence identity of 81%, indicating thatSjNPYR-like was closely related toObNPY2R phylogenetically and evolutionarily,consistent with the evolutionary relationship of these two species. And it also clustered with the functionally identified NPFR ofD.melanogaster,B.mori, andA.gambiae, suggesting its functional relationship with NPFRs.
GPCRs are described as seven-transmembrane receptors because they pass through the cell membrane seven times, and are located in the plasma membrane to sense molecules outside the cell and activate signal transduction pathways inside (Trzaskowski et al.,2012). The cell surface localization property ofSjNPYR-like is crucial for its functional activity. To further assess the characteristics ofSjNPYR-like as a transmembrane receptor, EGFP was fused to the C-terminus ofSjNPYR-like and expressed in HEK293 cells. The expression of this recptor on cell surface was demonstrated by confocal microscopy (Fig.5).With the putative 7TM domain structure ofSjNPYR-like from structure modeling (Fig.3), this result suggested that the cell surface localization property ofSjNPYR-like was consistent with functioning as a transmembrane G protein-coupled receptor. The interaction between this receptor and its ligand should be investigated in future to clarify the signaling pathway and the details of the cellular responses.
NPYRs are distributed ubiquitously in the central nervous system and peripheral tissues to mediate diverse physiological functions (Silva et al., 2005;Hausman et al., 2008; Pezeshki et al., 2012; Shi et al.,2012). The gene expression ofSjNPYR-likein various tissues was demonstrated here, and qRT-PCR analysis indicated thatSjNPYR-likemRNA was detected at high levels in tissues that are known to be involved in developmental and reproductive processes, such as the brain, liver, and ovary (Fig.7). NPY is known to play a key role in promoting food intake and energy budgeting in vertebrates (Frankish et al., 1995; White and Martin, 1997). However, there is evidence suggesting that NPY is involved in inhibiting the main energy-consuming processes in invertebrates,such as reproduction and growth (de Jong-Brink et al., 2001). The regulatory roles of NPYs in different species may be inconsistent. Further investigation on the expression profi le ofSjNPYR-likein brain, liver,and ovary during the growth and maturation process was conducted to evaluate its potential regulatory functions. The expression of NPYR in theS.japonicadevelopmental process indicated that the extremely low level ofSjNPYR-likemRNA in the ovary during period IV coincided with the rapid weight increase of the ovary during this stage. The high level ofSjNPYR-likemRNA in all brain and liver samples coincided with the persistent increase in body mass during the developmental process (Fig.6). These results suggested thatSjNPYR-like potentially plays a role in the inhibition of gonadal development and control of growth. However, more evidence is needed from protein quantifi cation and analysis of this receptor and its ligand (NPY/NPF) in these tissues to reveal subtle functional roles ofSjNPYR-like.
5 CONCLUSION
A full-length cDNA of the NPYR-like ofS.japonicawas obtained using RACE technology,and characteristics ofSjNPYR-like were investigated.The predictedSjNPYR-like protein showed a high degree ofidentity with the putative NPY2R protein sequence ofO.bimaculoides. TheSjNPYR-like transmembrane character was demonstrated using the HEK293 cell line and a recombinant plasmid encoding aSjNPYR-like-EGFP fusion protein. The observedSjNPYR-liketranscription profi le in multiple tissues suggested thatSjNPYR-like has diverse functions.Moreover,SjNPYR-likeexpression in the brain, liver,and ovary during the female gonadal developmental process indicated involvement ofSjNPYR-like in
control of growth and reproduction. Our results have led to a basic understanding of NPYR inS.japonica,and provide a foundation for further exploration of the signaling pathway and regulatory mechanism of this receptor. Further experiments should be conducted to clarify the interaction betweenSjNPYR-like and its ligand, cell signaling pathways involvingSjNPYR-like, and the fundamental physiological functions of this receptor.
猜你喜欢
杂志排行
Journal of Oceanology and Limnology的其它文章
- Editorial Statement
- The post-larval and juvenile fi sh assemblage in the Sukhothai Floodplain, Thailand*
- Effects of probiotic on microf l oral structure of live feed used in larval breeding of turbotScophthalmus maximus*
- Comparison ofintestinal microbiota and activities of digestive and immune-related enzymes of sea cucumberApostichopus japonicusin two habitats*
- Otolith shape analysis for stock discrimination of twoCollichthysgenus croaker (Pieces: Sciaenidae,) from the northern Chinese coast*
- The impact of spatial autocorrelation on CPUE standardization between two different fi sheries*