ldentification of QTLs associated with cadmium concentration in rice grains
2018-07-09HUDaweiSHENGZhonghuaLlQianlongCHENWeiWElXiangjinXlELihongJlAOGuiaiSHAOGaonengWANGJianlongTANGShaoqingHUPeisong
HU Da-wei, SHENG Zhong-hua, Ll Qian-long, CHEN Wei, WEl Xiang-jin, XlE Li-hong, JlAO Guiai, SHAO Gao-neng, WANG Jian-long, TANG Shao-qing, HU Pei-song,
1 State Key Laboratory of Rice Biology, Ministry of Science and Technology/Key Laboratory of Rice Biology and Breeding, Ministry of Agriculture/China National Rice Research Institute, Hangzhou 310006, P.R.China
2 College of Agronomy, Hunan Agricultural University, Changsha 410128, P.R.China
1. lntroduction
Cadmium (Cd) is a highly toxic heavy metal that enters the human food chain through crops planted on Cdcontaminated soil. Chronic intake of Cd-contaminated food can cause several serious diseases, such as renal dysfunction and cancer (Nawrot et al. 2006; Satarug et al.2011; Åkesson et al. 2014). A famous case of Cd toxicity is ‘itai-itai disease’, which occurred in the middle of the last century in Japan (Horiguchi et al. 1994). ‘Itai-itai disease’was caused by the long-term ingestion of Cd-contaminated rice and drinking water and led to the death of hundreds of local residents. Because rice (Oryza sativa) is a staple food for over half of the global population and a major dietary source in Asia (Yan et al. 2013; Huang et al. 2015),it is of immense importance to reduce Cd accumulation in rice grains, especially for people residing in areas where the arable land is seriously contaminated by Cd, such as partial arable land in southern China (Zhao et al. 2015; Jha and Bohra 2016).
In theory, two strategies have been proposed to reduce Cd accumulation in rice grains (Zhao et al. 2015). One strategy is to decrease Cd availability in the soil by applying soil dressing, applying alkaline amendments, and managing water (Ishikawa et al. 2005a). The other strategy is so-called phytoextraction, namely, removing Cd from soil by planting hyperaccumulators (Murakami et al. 2007). However,these techniques are impractical due to their high cost and low efficiency. Thus, breeding cultivars with low grain Cd concentrations is the only viable approach.
A deep understanding of the mechanisms controlling Cd accumulation in rice grains and excavation of valuable gene resources are the basis for developing a low-Cdconcentration rice cultivar. Recent studies have revealed that three major processes are responsible for Cd transport from the soil to the rice grain. These include root uptake and xylem loading, redirection of transport through intervascular transfer at nodes and remobilization from the leaf blade via phloem (Uraguchi and Fujiwara 2013). Using reverse genetics methods and QTL mapping, a number of genes involved in mediating these processes have been cloned. The manganese (Mn) transporter OsNramp5 is expressed on the distal side of the plasma membrane of the exodermis and endodermis, and compared with wild type, the osnramp5 mutant lost more than 90% of the Cd absorption (Sasaki et al. 2012). OsHMA3 is another Cd transporter functioning in vacuolar sequestration of Cd in root cells (Ueno et al. 2010; Miyadate et al. 2011;Takahashi et al. 2012a; Sasaki et al. 2014; Yan et al. 2016).Silencing OsHMA3 resulted in increased Cd translocation from roots to shoots, whereas overexpression produced the opposite effect (Ueno et al. 2010; Sasaki et al. 2014).OsHMA2 is homologous to OsHMA3 and is also mainly expressed in roots. The oshma2 mutant had lower Cd and Zn concentrations in reproductive organs (Satoh-Nagasawa et al. 2012; Takahashi et al. 2012b). Unlike the three transporters described above, OsLCT1 is expressed in phloem parenchyma cells of nodal vascular bundles,especially in diffuse vascular bundles, and OsLCT1 acts as an efflux-type transporter to facilitate Cd loading to the sieve tube by supplying Cd from the parenchyma cells(Uraguchi et al. 2011). Knock down of OsLCT1 reduced the Cd concentration in rice grains by half (Uraguchi et al.2011). In addition to these examples, OsIRT1 (Nakanishi et al. 2006), OsIRT2 (Nakanishi et al. 2006), OsHMA9 (Lee et al. 2007), OsNramp1 (Takahashi et al. 2011; Tiwari et al.2014), and OsLCD (Shimo et al. 2011) have been found to play a crucial role in Cd translocation in rice. These genes provide a theoretical basis and technical support for developing rice varieties with low Cd accumulation by using marker-assistance selection (MAS).
Quantitative trait locus (QTL) mapping is a powerful tool for studying complex agronomic traits controlled by multiple genes, and it has been successfully applied to detect loci controlling Cd accumulation in rice. Ishikawa et al. (2005b)first reported that three putative QTLs controlling Cd concentration in brown rice were located on chromosomes 3, 6, and 8 using 39 CSSLs (chromosome segment substitution lines). Ueno et al. (2009) identified a major QTL controlling shoot Cd concentration on chromosome 11 using an F2population. Abe et al. (2013) detected a major QTL,qlGCd3 controlling Cd content in rice grains on the long arm of chromosome 3 using 46 CSSLs. Although a number of QTLs associated with Cd accumulation and tolerance in rice have been identified, it is still necessary to mine more QTLs and genes that can be directly used to breed low-Cdaccumulation varieties by MAS to deal with the increasingly serious problem of Cd contamination in rice.
2. Materials and methods
2.1. Plant materials
A DH population consisting of 101 lines was used to map QTLs for Cd concentration in brown and milled rice. The male parent D50 is a tropical japonica cultivar with a relatively high Cd concentration in grain and female parent Zhongjiazao 17 (YK17) is an early-season indica cultivar with low grain Cd concentration.
2.2. Pot and field trials
Pot trials were conducted in Hangzhou City (HZ), Zhejiang Province, China, in 2015 and 2016, and the total soil Cd concentration was 6.17 and 2.98 mg kg–1, respectively.There were no differences in the cultivation procedures between the two pot trials except for soil Cd concentration.Seeds of the DH population and parents were sown on the 21st May and seedlings were transplanted into plastic pots containing 9 kg soil on the 15th June. Two seedlings from different lines were planted in each pot. Compound fertilizer (3 g) was applied as a basal dose, and an additional 1 g urea was top-dressed at the tillering stage. The water consumption rate in each pot was much different due to the difference in growth rate and biomass of the different lines.So, we watered once every 2 days, each time adding water until the water level was 2–3 cm above the soil surface. The soil was kept flooded until the grain ripened in October so as to eliminate experimental error due to the difference in water consumption of each pot.
Field trials were conducted in HZ (30°N, 120°E) in 2015 and 2016 and in Hainan Province (HN) (18.4°N, 109°E) in 2015, and the total soil Cd concentrations were 0.56, 0.45,and 0.16 mg kg–1, respectively. In the 2015 and 2016 HZ field trials, seeds were sown on 21st May and seedlings were transplanted on 15th June, while in the 2015 HN field trial,seeds were sown on 15th November and seedlings were transplanted on 28th December. Each line was planted in a 5-row plot with 8 plants per row with 15 cm×21 cm spacing under a randomized complete block design with three replications. Irrigation, fertilization, and other management measures followed normal field practices.
2.3. Cd determination
After harvest, the grains of DH lines and their parents were ground into brown rice and milled rice, and the samples were further ground into a fine powder for Cd detection.The brown rice and milled rice powders were dried at 80°C for 24 h. After drying, 0.25 g powder was weighed and digested with 5 mL 65% HNO3solution according to a microwave digestion protocol (MARS Xpress, CEM,America). The digested solution was heated at 150°C for acid-driving and when the volume of solution had evaporated to 1 mL, Milli-Q water was added to a final volume of 20 mL.The Cd concentrations in brown rice (CCBR) and milled rice (CCMR) were determined using an atomic absorption spectrophotometer (AA-6800, Shimadzu, Japan), and certified standard material was used as a control to ensure the precision of the analytical procedure.
2.4. Linkage map construction and QTL analysis
A linkage map of the DH population was constructed using 170 SSRs (simple sequence repeats) polymorphic markers evenly distributed on 12 chromosomes. MAPMAKER/EXP version 3.0 (Lincoln et al. 1993) was used to construct the linkage map, and recombination rate was converted into genetic distance (cM) using the Kosambi function. QTLs affecting Cd concentration in brown rice and milled rice were mapped using the composite interval mapping (CIM)function in Windows QTL Cartographer version 2.5 (Wang et al. 2007), and a LOD value of 2.5 was used as the threshold for detection of putative QTLs. QTLs with epistatic effects and environment interaction effects were analyzed using the mixed-model-based composite interval mapping(MCIM) function in QTL Network-2.1 (Yang et al. 2008).
2.5. Statistical analysis
All data were analyzed using SPSS 19.0 and Excel 2016.
3. Results
3.1. Phenotypic variation in Cd concentration in the DH population and parents
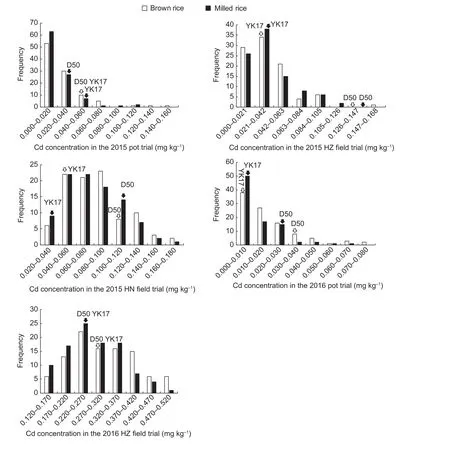
CCBR and CCMR for the DH population and the parents are shown in Table 1 and Fig. 1. Over five trials, thevariation in CCBR and CCMR in the DH population was continuous and wide, and both traits exhibited twodirection transgressive segregation except for CCMR in the 2015 HZ field trial. These results indicate that CCBR and CCMR are quantitatively inherited traits controlled by multiple genes.
60年前的兰州市西固区,是一片荒凉之地,“地上不长草,风吹石头跑”。老一辈“两兰人”硬是依靠人拉肩扛在滩涂地上建起了炼化生产企业,生产出了新中国第一批汽煤柴油和丁苯橡胶。新一代兰州石化人牢记使命,砥砺前进,续写了新的辉煌,实现了炼油化工从无到有、从小到大、由弱到强的巨变。
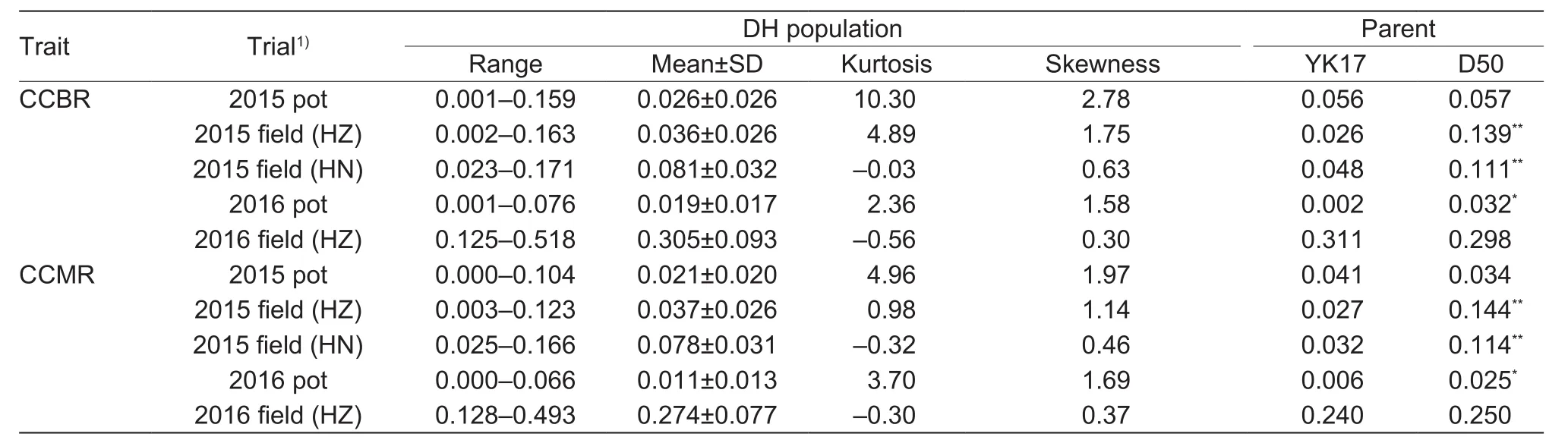
Table 1 Cd concentration in brown rice and milled rice in the double haploid (DH) population and parents (mg kg–1)
In comparison to D50, YK17 showed lower CCBR and CCMR over five trials except the CCBR in 2016 HZ field trial and CCMR in 2015 pot trail.
3.2. Correlation analysis of CCBR and CCMR
Correlation analysis revealed a positive and highly significant correlation between CCBR and CCMR in each trial (Fig. 2),with correlation coefficients ranging from 0.661 to 0.886.
3.3. QTL mapping of CCBR and CCMR in the DH population
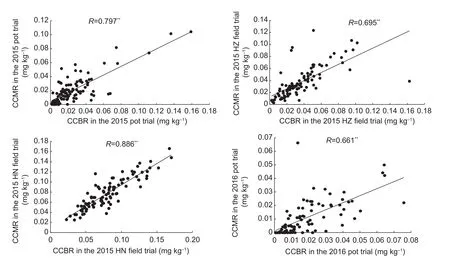
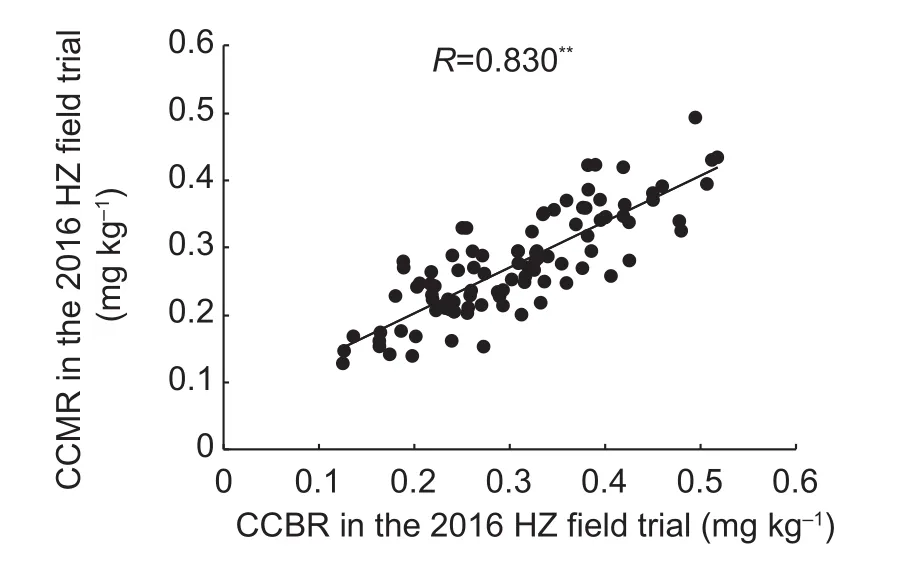
Fig. 2 Correlation of Cd concentration in brown rice (CCBR) and Cd concentration in milled rice (CCMR) in the double haploid(DH) population. HZ, Hanzhou City, Zhejiang Province, China;HN, Hainan Province, China. ** indicates significance at the 0.01 probability level.
QTLs controlling CCBR and CCMR in the DH population are summarized in Table 2, and the chromosomal locations of these QTLs are shown in Fig. 3. For CCBR, a total of 18 QTLs were detected in five trials. These QTLs were distributed on all chromosomes except for chromosomes 6 and 8, and single QTLs explained 6.7–24.1% of the phenotypic variance. Two pairs of QTLs (qCCBR2-1 and qCCBR2-2) and (qCCBR9-1 and qCCBR9-2) that mapped to the same chromosomal interval were detected in multiple trials; the average proportion of phenotypic variance explained by each pair was 16.3 and 21.2%, respectively,and the alleles increasing CCBR were contributed by YK17 and D50, respectively. Another pair of QTLs with overlapping marker intervals, qCCBR4-1 and qCCBR4-2, are unlikely to be the same QTL because of the large distance between the peak positions, even though the additive effects of both come from YK17.
For CCMR, a total of 14 QTLs were detected in four trials, and no QTL was identified in the 2016 pot trial. These QTLs were distributed on chromosomes 1, 2, 3, 4, 5, 7,and 11, and single QTL accounted for 6.6–20.3% of the phenotypic variance. qCCMR5-1 and qCCMR5-2, which were located in approximately the same interval, were the only QTLs detected in multiple trials. For both QTLs, the peak positions were located in adjacent marker intervals,and the D50 allele increased CCMR.
By comparing the mapping results for CCBR and CCMR,it was found that eight pairs of QTLs for CCBR and CCMR(qCCBR2-2 and qCCMR2-2, qCCBR3 and qCCMR3,qCCBR4-2 and qCCMR4-1, qCCBR4-3 and qCCMR4-2,qCCBR4-4 and qCCMR4-3, qCCBR5 and qCCMR5-2,qCCBR7 and qCCMR7, qCCBR11-1 and qCCMR11-2)co-localized on chromosomes 2, 3, 4, 5, 7 and 11, and that the peak positions of each pair were very close or identical except for qCCBR4-2 and qCCMR4-1. Furthermore, among the eight pairs of QTLs, the additive effect of seven comes from the YK17 allele, indicating that CCBR and CCMR are controlled by similar loci.
Four QTLs (qCCBR2, qCCMR2, qCCMR4, and qCCMR5) with significant additive×environment interaction were detected by joint analysis of CCBR and CCMR in five trials (Table 3). In the 2016 HZ field trial, the interaction effect was significant to extremely significant for four QTLs.The phenotypic contribution rate was relatively high, which suggests these QTL effects were greatly influenced by theenvironmental conditions. In addition, two pairs of bi-allelic epistatic interactions for CCMR were detected (Table 4),one between QTLs located in the middle of chromosomes 2 and 11, which could explain 4.3% of the phenotypic variance, and the other between QTLs located in the middle of chromosome 8 and the bottom of chromosome 10, which could explain 2.8% of the phenotypic variance.
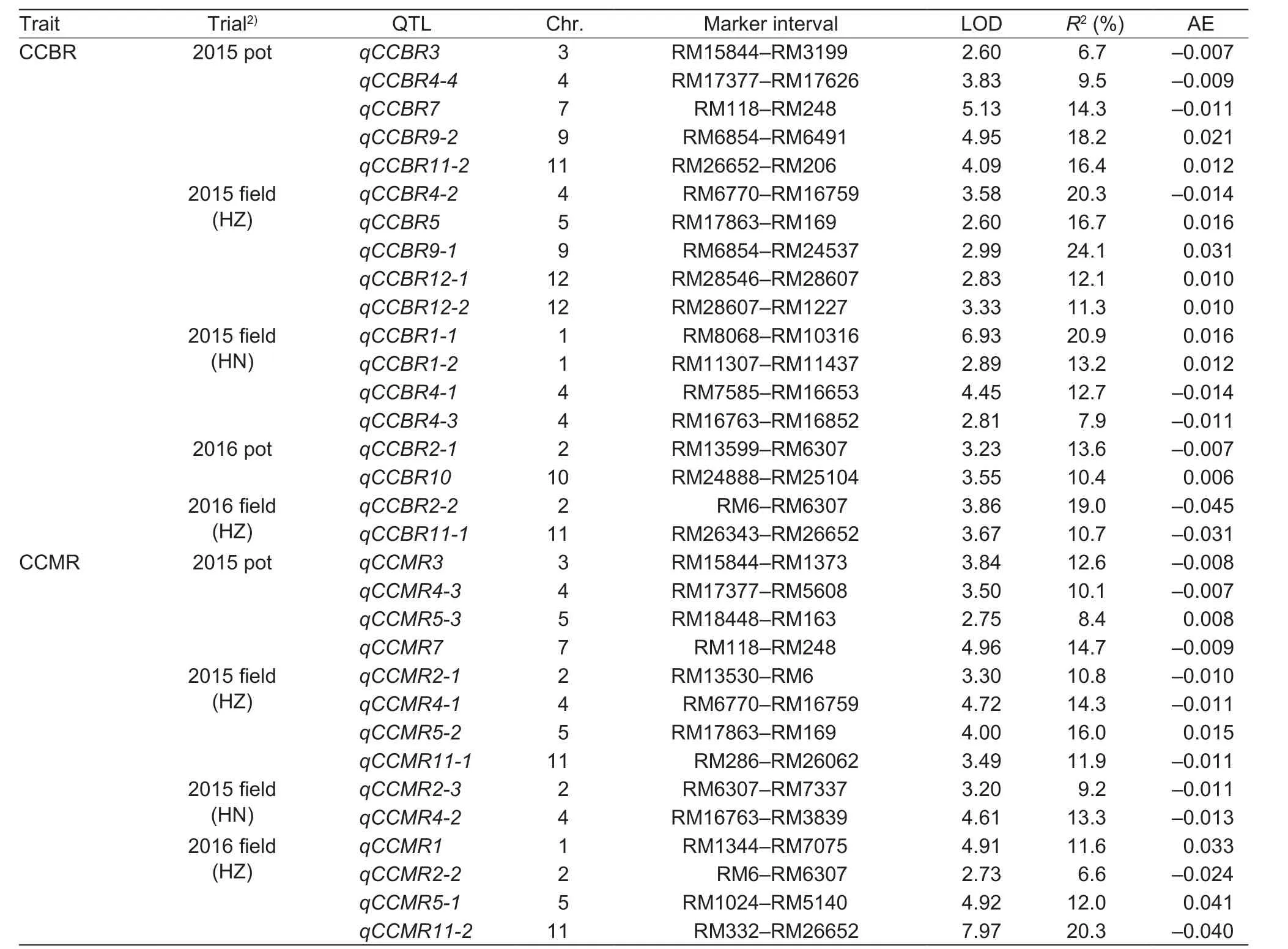
Table 2 QTLs controlling Cd concentration in brown rice (CCBR) and Cd concentration in milled rice (CCMR) in the double haploid(DH) population under five different environmental conditions1)
4. Discussion
4.1. CCBR and CCMR mostly share the same genetic mechanism in the Cd
In comparison to brown rice, milled rice grasps most of the market share and the consumer’s table, so it is more practical to study the mechanisms of Cd accumulation in milled rice. However, only a few earlier studies explored this area of research. In the present study, we initially detected 14 QTLs controlling CCMR. By comparing these QTLs with those mapped for CCBR, we found that eight pairs of QTLs associated with both CCBR and CCMR were located in the same chromosomal regions. Furthermore, the peak positions of these QTL pairs were very close or identical,and the directions of the additive effects were the same.Correlation analysis suggested that CCBR and CCMR were highly and positively correlated in each trial. Based on these results, we conclude that there are similarities in the genetic mechanism controlling CCBR and CCMR.Similar conclusions were also drawn in previous studies where QTLs were mapped for protein content in brown and milled rice (Zhong et al. 2007; Yang et al. 2012). In these studies, BRPC (protein content in brown rice) was highly correlated with MRPC (protein content in milled rice) in the same planting environment. It should be noted that there was also a large number of QTLs for CCBR and CCMR that didn’t co-localize, which may be due to the uneven distribution of Cd in rice, and more research is required to uncover this mechanism.
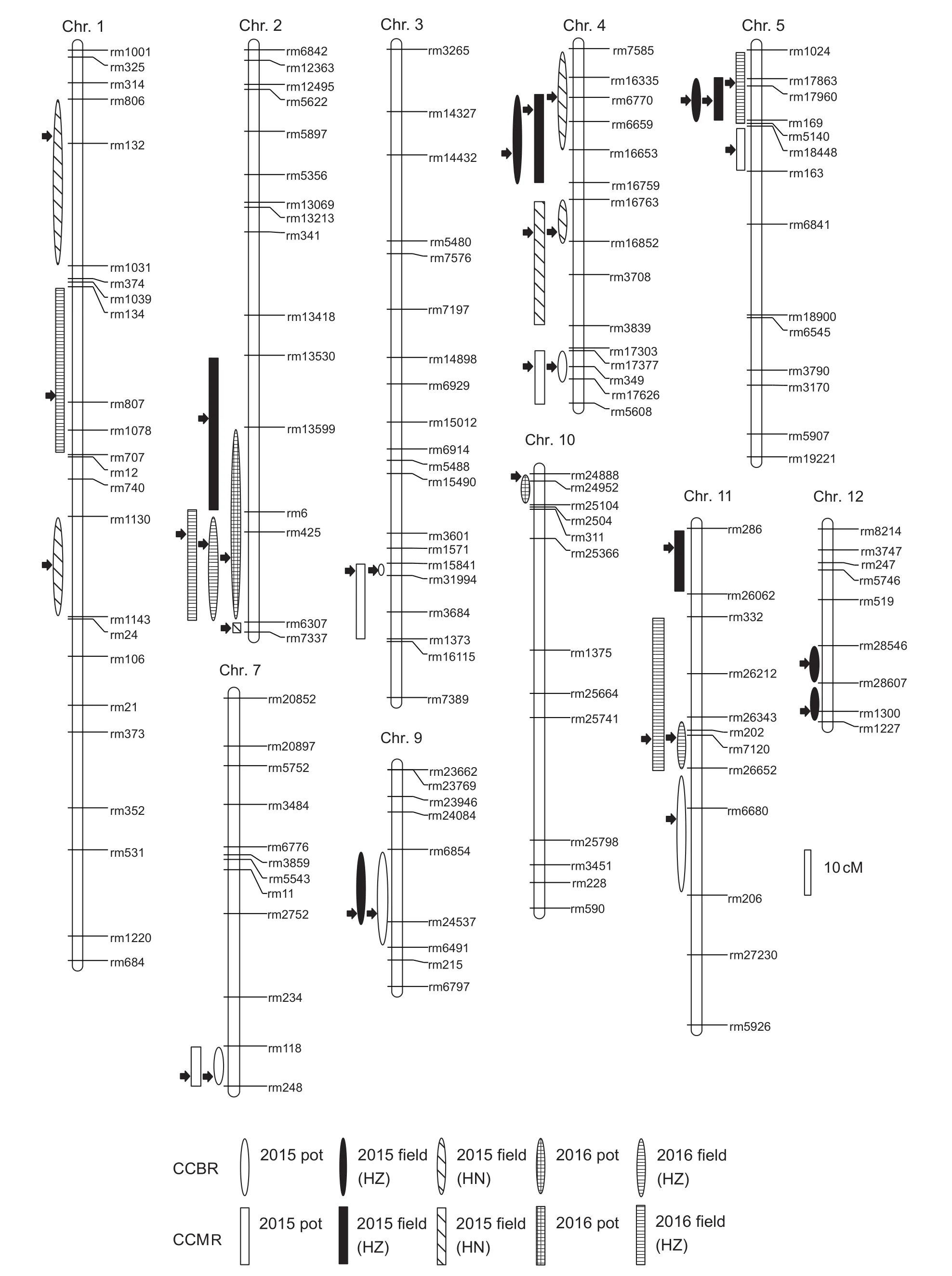
Fig. 3 Chromosomal distribution of QTLs for Cd concentration in brown rice (CCBR) and Cd concentration in milled rice (CCMR)under five environmental conditions. The black arrows indicate the peak positions of putative QTLs. HZ, Hangzhou City, Zhejiang Province, China; HN, Hainan Province, China.
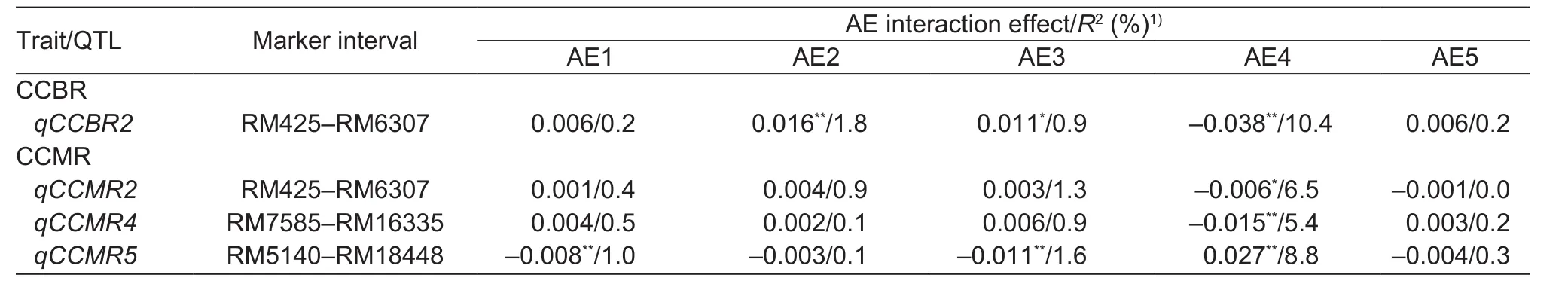
Table 3 QTLs for Cd concentration in brown rice (CCBR) and Cd concentration in milled rice (CCMR) in the double haploid (DH)population with significant additive×environment interactions

Table 4 Epistatic interactions between Cd concentration in milled rice (CCMR) QTLs in the double haploid (DH) population1)
4.2. The expected phenotypic variation of Cd concentration in rice grain may be caused by different cultivation practices
The Cd concentrations in D50 in each of the five pot and field trials were mostly higher than those in YK17 with a few exceptions. The Cd concentration of the two parents in the 2015 HZ pot trial and the 2016 HZ field trial were almost the same. The total soil Cd concentration in the pot trial was much higher than that in the field trial, but in general Cd concentration in the DH population and the two parents under some field trials was not lower than those in pot trials. Even the Cd concentration in grain in the 2016 HZ field trial was several times higher than that in the pot trials; this unexpected finding may be associated with variation in available Cd in the soil during different trials due to differences in soil physicochemical properties and our cultivation practices. It is well understood that Cd uptake by rice varies greatly depending on soil redox potential.Cd bioavailability in soil increases under oxidative upland conditions owing to formation of soluble cadmium sulfate(CdSO4) and decreases under reductive flooded conditions because of the formation of the less-soluble cadmium sulfide(CdS). As a result, the rice grain Cd concentration was much lower under flooded conditions than that under upland conditions (Ishikawa et al. 2010). Thus, due to their effect on soil redox state, water and fertilizer management practices cause phenotypic variation in grain Cd concentration.
4.3. Some repeated detected and novel QTLs of rice grain Cd concentration are worthwhile to further cloning
Based on comparisons of our QTLs to those identified by other researchers, some QTLs are likely identical. Abe et al. (2013) identified a QTL (qlGCd3) associated with low grain Cd content on chromosome 3, which is located in the same position as qCCBR3 and qCCMR3. Yan et al. (2013)reported three QTLs for grain Cd concentration. One of these QTLs (gcc11) was located in the same chromosomal region as qCCMR11-1. Moreover, some QTLs detected in our study overlap with cloned Cd-related genes. For example, qCCMR5-1 was mapped to the interval RM1024–RM169, which contains OsMTP1 (Yuan et al. 2012), and the mapping interval of qCCBR10 contains OsPCR1 (Song et al. 2015). In addition, three pairs of novel and important loci for grain Cd concentration on chromosomes 2, 4, and 9, were detected in multiple trials in our study, qCCBR2-2 and qCCMR2-2, could explain 19.0 and 6.6% of the phenotypic variance, respectively with background from YK17. The qCCBR4-2 and qCCMR4-1 QTLs could explain 20.3 and 14.3% of the phenotypic variance, respectively,and these alleles also came from YK17. The qCCBR9-1 and qCCBR9-2 QTLs could explain an average of 21.2%of the phenotypic variance, and the favorable allele was contributed by D50. These loci have a high phenotypic contribution rate and good stability; therefore, it is worthwhile to perform fine mapping and even cloning by constructing a secondary segregating population, which is currently under way in our research group.
4.4. The QTLs of CCRR and CCMR could be affected by environmental factors and epistatic effection
In this study, 18 QTLs for CCBR and 14 QTLs for CCMR were identified, but only a few pairs of QTLs for each trait could be detected in multiple trials, and no QTL was detected in three or more trials. This indicates that these QTLs are differentially expressed under different environmental conditions. The inheritance of quantitative traits is easily influenced by several environmental factors such as climate and cultivation measures; therefore, the results of QTL mapping under multiple planting environments will be different (Lu et al. 1997; Zhuang et al. 1997; Xing et al.2002). Four QTLs with significant additive×environment interaction for CCBR and CCMR were detected, which also illustrates the environmental influence on QTL expression.In addition to environmental factors, the interaction between QTLs or genes could also affect the expression of QTLs(Xing et al. 2002; Zhong et al. 2007; Yang et al. 2016).Two pairs of epistatic QTLs for CCMR were detected in this study, which partially explains why only one pair of CCMR QTLs was detected in multiple trials and also reveals that the regulatory mechanism controlling Cd concentration in grain is very complex.
5. Conclusion
In this study, a DH population derived from Zhongjiazao 17(YK17)×D50 was used to map QTLs associated with Cd concentration in brown rice and milled rice. There was a positive and highly significant correlation between CCBR and CCMR. Continuous and wide variation in CCBR and CCMR was observed among the DH population. A total of 18 QTLs for CCBR and 14 QTLs for CCMR were identified under five different pot and field trials, and two pairs of QTLs for CCBR (qCCBR2-1 and qCCBR2-2, qCCBR9-1 and qCCBR9-2) and one pair of QTLs for CCMR (qCCMR5-1 and qCCMR5-2) were detected in multiple trials. Eight pairs of QTLs for CCBR and CCMR (qCCBR2-2 and qCCMR2-2, qCCBR3 and qCCMR3, qCCBR4-2 and qCCMR4-1, qCCBR4-3 and qCCMR4-2, qCCBR4-4 and qCCMR4-3, qCCBR5 and qCCMR5-2, qCCBR7 and qCCMR7, qCCBR11-1 and qCCMR11-2) co-localized on chromosomes 2, 3, 4, 5, 7, and 11, respectively. In addition, three pairs of novel and important loci (qCCBR2-2 and qCCMR2-2, qCCBR4-2 and qCCMR4-1, qCCBR9-1 and qCCBR9-2, located on chromosomes 2, 4, and 9,respectively) with high contributions to Cd concentration in grain, were repeatedly detected in our study. These results could facilitate the cloning of Cd concentration-associated genes in rice and MAS for breeding rice varieties with low Cd accumulation in grain.
Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (31501285),the Zhejiang Science and Technology Projects, China(2015C32045), the National Key Research and Development Program of China (2016YFD0101801), and the National S&T Major Project, China (2016ZX08001006).
Abe T, Nonoue Y, Ono N, Omoteno M, Kuramata M, Fukuoka S, Yamamoto T, Yano M, Ishikawa S. 2013. Detection of QTLs to reduce cadmium content in rice grains using LAC23/Koshihikari chromosome segment substitution lines.Breeding Science, 63, 284–291.
Åkesson A, Barregard L, Bergdahl I A, Nordberg G F,Nordberg M, Skerfving S. 2014. Non-renal effects and the risk assessment of environmental cadmium exposure.Environmental Health Perspectives, 122, 431–438.
Horiguchi H, Teranishi H, Niiya K, Aoshima K, Katoh T,Sakuragawa N, Kasuya M. 1994. Hypoproduction of erythropoietin contributes to anemia in chronic cadmium intoxication: Clinical study on itai-itai disease in Japan.Archives of Toxicology, 68, 632–636.
Huang Y, Sun C X, Min J, Chen Y L, Tong C, Bao J S. 2015.Association mapping of quantitative trait loci for mineral element contents in whole grain rice (Oryza sativa L.).Journal of Agricultural and Food Chemistry, 63, 10885–10892.
Ishikawa S, Abe T, Kuramata M, Yamaguchi M, Ando T,Yamamoto T, Yano M. 2010. A major quantitative trait locus for increasing cadmium-specific concentration in rice grain is located on the short arm of chromosome 7. Journal of Experimental Botany, 61, 923–934.
Ishikawa S, Ae N, Sugiyama M, Murakami M, Arao T. 2005a.Genotypic variation in shoot cadmium concentration in rice and soybean in soil with different levels of cadmium contamination. Soil Science and Plant Nutrition, 51,101–108.
Ishikawa S, Ae N, Yano M. 2005b. Chromosomal regions with quantitative trait loci controlling cadmium concentration in brown rice (Oryza sativa). New Phytologist, 168, 345–350.
Jha U C, Bohra A. 2016. Genomics enabled breeding approaches for improving cadmium stress tolerance in plants. Euphytica, 208, 1–31.
Lee S, Kim Y Y, Lee Y, An G. 2007. Rice P-1B-type heavymetal ATPase, OsHMA9, is a metal efflux protein. Plant Physiology, 145, 831–842.
Lincoln S E, Daly M J, Lander E S. 1993. Constructing Genetic Linkage Maps with Mapmaker/Exp Version 3.0: A Tutorial and Reference Manual. A Whitehead Institute for Biomedical Research Technical Report. 3rd ed. Whitehead Institute for Biometrical Research, Cambridge, Massachusetts.
Lu C F, Shen L S, Tan Z B, Xu Y B, He P, Chen Y, Zhu L H.1997. Comparative mapping of QTLs for agronomic traits of rice across environments by using a doubled haploid population. Theoretical and Applied Genetics, 94, 145–150.
Miyadate H, Adachi S, Hiraizumi A, Tezuka K, Nakazawa N,Kawamoto T, Katou K, Kodama I, Sakurai K, Takahashi H, Satoh-Nagasawa N, Watanabe A, Fujimura T, Akagi H. 2011. OsHMA3, a P-1B-type of ATPase affects root-toshoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytologist, 189, 190–199.
Murakami M, Ae N, Ishikawa S. 2007. Phytoextraction of cadmium by rice (Oryza sativa L.), soybean (Glycine max (L.) Merr.), and maize (Zea mays L.). Environmental Pollution, 145, 96–103.
Nakanishi H, Ogawa I, Ishimaru Y, Mori S, Nishizawa N K. 2006.Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+transporters OsIRT1 and OsIRT2 in rice. Soil Science and Plant Nutrition, 52, 464–469.
Nawrot T, Plusquin M, Hogervorst J, Roels H A, Celis H,Thijs L, Vangronsveld J, Hecke E V, Staessen J A. 2006.Environmental exposure to cadmium and risk of cancer:A prospective population-based study. Lancet Oncology,7, 119–126.
Sasaki A, Yamaji N, Ma J F. 2014. Overexpression of OsHMA3 enhances Cd tolerance and expression of Zn transporter genes in rice. Journal of Experimental Botany, 65, 6013–6021.
Sasaki A, Yamaji N, Yokosho K, Ma J F. 2012. Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. The Plant Cell, 24, 2155–2167.
Satarug S, Garrett S H, Sens M A, Sens D A. 2011. Cadmium,environmental exposure, and health outcomes. Ciência &Saúde Coletiva, 16, 2587–2602. (in Portuguese)
Satoh-Nagasawa N, Mori M, Nakazawa N, Kawamoto T, Nagato Y, Sakurai K, Takahashi H, Watanabe A, Akagi H. 2012.Mutations in rice (Oryza sativa) Heavy Metal ATPase2(OsHMA2) restrict the translocation of zinc and cadmium.Plant and Cell Physiology, 53, 213–224.
Shimo H, Ishimaru Y, An G, Yamakawa T, Nakanishi H,Nishizawa N K. 2011. Low cadmium (LCD), a novel gene related to cadmium tolerance and accumulation in rice.Journal of Experimental Botany, 62, 5727–5734.
Song W Y, Lee H S, Jin S R, Ko D, Martinoia E, Lee Y, An G,Ahn S N. 2015. Rice PCR1 influences grain weight and Zn accumulation in grains. Plant, Cell & Environment, 38,2327–2339.
Takahashi R, Bashir K, Ishimaru Y, Nishizawa N K, Nakanishi H. 2012a. The role of heavy-metal ATPases, HMAs, in zinc and cadmium transport in rice. Plant Signaling and Behavior, 7, 1605–1607.
Takahashi R, Ishimaru Y, Senoura T, Shimo H, Ishikawa S, Arao T, Nakanishi H, Nishizawa N K. 2011. The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. Journal of Experimental Botany, 62, 4843–4850.
Takahashi R, Ishimaru Y, Shimo H, Ogo Y, Senoura T,Nishizawa N K, Nakanishi H. 2012b. The OsHMA2 transporter is involved in root-to-shoot translocation of Zn and Cd in rice. Plant, Cell & Environment, 35, 1948–1957.
Tiwari M, Sharma D, Dwivedi S, Singh M, Tripathi R D, Trivedi P K. 2014. Expression in Arabidopsis and cellular localization reveal involvement of rice NRAMP, OsNRAMP1, in arsenic transport and tolerance. Plant, Cell & Environment, 37,140–152.
Ueno D, Kono I, Yokosho K, Ando T, Yano M, Ma J F. 2009.A major quantitative trait locus controlling cadmium translocation in rice (Oryza sativa). New Phytologist, 182,644–654.
Ueno D, Yamaji N, Kono I, Huang C F, Ando T, Yano M, Ma J F. 2010. Gene limiting cadmium accumulation in rice.Proceedings of the National Academy of Sciences of the United States of America, 107, 16500–16505.
Uraguchi S, Fujiwara T. 2013. Rice breaks ground for cadmiumfree cereals. Current Opinion in Plant Biology, 16, 328–334.
Uraguchi S, Kamiya T, Sakamoto T, Kasai K, Sato Y, Nagamura Y, Yoshida A, Kyozuka J, Ishikawa S, Fujiwara T. 2011.Low-affinity cation transporter (OsLCT1) regulates cadmium transport into rice grains. Proceedings of the National Academy of Sciences of the United States of America,108, 20959–20964.
Wang S, Basten C J, Zeng Z B. 2007. Windows QTL Cartographer Version 2.5. North Carolina State University,USA.
Xing Y Z, Tan Y F, Hua J P, Sun X L, Xu C G, Zhang Q. 2002.Characterization of the main effects, epistatic effects and their environmental interactions of QTLs on the genetic basis of yield traits in rice. Theoretical and Applied Genetics,105, 248–257.
Yan J L, Wang P T, Wang P, Yang M, Lian X M, Tang Z, Huang C F, Salt D E, Zhao F J. 2016. A loss-of-function allele of OsHMA3 associated with high cadmium accumulation in shoots and grain of japonica rice cultivars. Plant, Cell &Environment, 39, 1941–1954.
Yan Y F, Lestari P, Lee K J, Kim M Y, Lee S H, Lee B W.2013. Identification of quantitative trait loci for cadmium accumulation and distribution in rice (Oryza sativa).Genome, 56, 227–232.
Yang J, Hu C C, Hu H, Yu R D, Xia Z, Ye X Z, Zhu J. 2008.QTLNetwork: Mapping and visualizing genetic architecure of complex traits in experimental populations. Bioinformatics,24, 721–723.
Yang L M, Sun J, Zhao H W, Wang J G, Liu H L, Zou D T. 2016.QTL analysis of heading date and yield traits in japonica rice under cold water stress in different years. Scientia Agricultura Sinica, 49, 3489–3503. (in Chinese)
Yang Y C, Ni D H, Song F S, Li L, Feng G, Li Z F, Yang J B.2012. Identification of QTL for protein content in brown and milled rice in two enironment. Chinese Journal of Rice Science, 26, 351–355. (in Chinese)
Yuan L Y, Yang S G, Liu B X, Zhang M, Wu K Q. 2012. Molecular characterization of a rice metal tolerance protein, OsMTP1.Plant Cell Reports, 31, 67–79.
Zhao F J, Ma Y B, Zhu Y G, Tang Z, McGrath S P. 2015. Soil Contamination in China: Current status and mitigation strategies. Environmental Science & Technology, 49,750–759.
Zhong M, Wang L Q, Luo L J, He Y Q. 2007. Comparison of quantitative trait loci controlling the protein content of brown and milled rice using a recombinant inbred line population.Molecular Plant Breeding, 5, 631–638.
Zhuang J Y, Lin H X, Lu J, Qian H R, Hittalmani S, Huang N,Zheng K L. 1997. Analysis of QTL×environment interaction for yield components and plant height in rice. Theoretical and Applied Genetics, 95, 799–808.
猜你喜欢
杂志排行
Journal of Integrative Agriculture的其它文章
- Characterisation of pH decline and meat color development of beef carcasses during the early postmortem period in a Chinese beef cattle abattoir
- Multi-mycotoxin exposure and risk assessments for Chinese consumption of nuts and dried fruits
- Effects of 1-methylcyclopropene and modified atmosphere packaging on fruit quality and superficial scald in Yali pears during storage
- Insertion site of FLAG on foot-and-mouth disease virus VP1 G-H loop affects immunogenicity of FLAG
- Update of Meat Standards Australia and the cuts based grading scheme for beef and sheepmeat
- Light shading improves the yield and quality of seed in oil-seed peony (Paeonia ostii Feng Dan)
