Simultaneous Biological Removal of p-Cresol, Sulfide and Nitrate by Denitrification
2018-01-19LiuChunshuangLiWenfeiZhaoDongfengLiWeiLiXuechenZhaoChaochengLiuFang
Liu Chunshuang; Li Wenfei; Zhao Dongfeng; Li Wei; Li Xuechen; Zhao Chaocheng; Liu Fang
(College of Chemical Engineering, China University of Petroleum, Qingdao 266580, China)
1 Introduction
Petrochemical or chemical industrial wastewaters, such as sour water, usually contain high levels of cresols, sulfides and nitrogenous compounds (i.e. ammonia and nitrates)[1].Cresols were classified as the pollutant of group C(possible human carcinogen) by the US Environmental Protection Agency[2]. p-Cresol has adverse effects on the central nervous system, cardiovascular system, lungs and kidneys and can cause central nervous system depression even at very low concentration[3]. Sulfide is a toxic,odorous, and corrosive substance in acidic environments.Ammonium radical contributes mainly to eutrophication of water bodies, besides the risks associated with its toxicity and offensive odor.
Biological removal of sulfur, nitrates, or phenolic compounds is a cost-effective and environmentally friendly process[4]. Beristain-Cardoso, et al.[5]achieved the simultaneous removal of nitrates, sulfides, and phenol in one reactor by the combination of autotrophic and heterotrophic bacteria provided with unlimited nitrate source. Beristain-Cardoso, et al. proposed that the oxidation of sulfide and phenol could be carried out under mixotrophic denitrification in the following way:sulfide was rapidly oxidized to elemental sulfur and then to sulfate, and after that phenol was oxidized to carbon dioxide. The maximum loading rates were 600 gS/(m3·d)for sulfide, 900 gN/(m3·d) for nitrate, and 450 gC/(m3·d)for phenol by granular sludge, as reported by Liu, et al.[6]However, up to now very scarce study was related with the simultaneous removal of cresols, sulfides and nitrates.Although, Francisco J. Cervantes[7]observed that the simultaneous removal of p-cresol, sulfides and nitrates could occur in the batch mode and the nitrite reduction was a rate-limiting step. The reactor performance in the continuous fl ow mode was not determined and the strains working in this process were not identified. Thereby, this study intended to investigate the performance of UASB reactor for simultaneous removal of nitrate, sulfide and p-cresol. The functional strains responsible for sulfide and p-cresol removal were identified.
2 Methods
2.1 Reactor and synthetic wastewater
The plexiglas up fl ow anaerobic sludge blanked (UASB)reactor, a modified version of Liu, et al.[8], was 50 mm in diameter and 75 cm in height, with a working volume of 1.0 L. The temperature in the reactor was kept at 29± 1 °C.The influent was introduced at the column bottom by aperistaltic pump. A gas-washing device at the column top collects the H2S gas generated during the experiment.Internal circulation from the sedimentation region to the column bottom forms a reflux ratio of 6.25:1.
Synthetic wastewater containing the sulfide,p-cresol,nitrate, bicarbonate, and trace elements was introduced into the reactor. The pH value of the synthetic wastewater was adjusted to 7.5. To investigate the performance of UASB reactor, the concentration of all compounds was doubled at a hydraulic retention time (HRT) of 12 h; then HRT was shortened from 12 h to 10 h, and further to 7 h. The detailed operation schemes of UASB reactor is described in Table 1. To investigate the effects of C/S ratio on the reactor performance, the influent concentration of sulfide and nitrate was maintained at 100 mgS/L and 100 mgN/L, respectively, in order to attain a S/N ratio of 1:1. Then four C/S ratios, viz. 0.75:1,1.0:1, 1.5:1, and 1.75:1, respectively, were tested in the present reactor by maintaining ap-cresol concentration of 75 mgC/L, 100 mgC/L, 150 mgC/L, and 175 mgC/L,respectively. The corresponding loading of sulfide and nitrate was 240 gS/m3d and 240 gN/m3d, respectively,while that ofp-cresol was 180 gC/(m3·d), 240 gC/(m3·d),360 gC/(m3·d) and 420 gC/(m3·d), respectively. The inoculated sludge was obtained from an anaerobic sludge thickener at the Nibuwan Municipal Wastewater Treatment Plant, Qingdao, China. The other details on reactor setup are available from the paper written by Liu,et al.[8]

Table 1 Experimental phases used during 39-day operation of the reactor
2.2 Chemical analysis
The concentrations of nitrate, nitrite, sulfate and thiosulfate in the collected liquor samples after filtration with a sieve mesh of 0.45 μm were measured by ion chromatography (ICS-3000; Dionex, USA). The sample separation and elution were performed using an IonOacAG4AAS4A-SC 4-mm analytical column operating with carbonate/bicarbonate eluent (1.8 mmol/L of Na2CO3/1.7 mmol/L of NaHCO3at a flowrate of 1 cm3/min) and sulfuric regeneration (25 mmol/L of H2SO4at a flowrate of 5 cm3/min). The sulfide concentration was determined using the methylene blue method[9]. The term “sulfide” in this study included all species in liquid (H2S(aq), HS-and S2-). The concentration of ammonium radical was analyzed[9].The pH value of the liquid samples was determined by a pH-meter (pH S-25;Shanghai, China). The SS and VSS of the sludge were measured according to the standard methods approved in China.
2.3 Microbial population
The sludge sample was collected from the EGSB reactor and immediately stored at -80 °C. Total genomic DNA of samples was extracted in duplicate using the PowerSoil DNA Isolation Kit (MoBio, Carlsbad, CA) according to the manufacturer’s instructions. The quality of the DNA extracted was examined by electrophoresis with 1% (w/v) of agarose gel and the concentration was measured with a UV-Vis spectrophotometer (NanoDrop 2000, USA). The V3-V4 region of the 16S rRNA gene was amplified using bacterial primers 338 F (5’-ACT CCT ACG GGA GGC AGC AG-3’) and 806 R (5’-GGA CTA CHV GGG TWT CTA AT-3’), with the reverse primer containing a 6-bp barcode used to tag each sample. The purified amplicon was quantified using a QuantiFluor-ST Fluorometer (Promega, USA), and then a composite sequencing library was constructed by combining equimolar ratios of amplicons from all samples. The resulting library for paired-end sequencing(2×250 bp) was analyzed on an Illumina Miseq platform(provided by the Majorbio Bio-Pharm Technology Co.,Ltd., Shanghai, China).
3 Results and Discussion
3.1 Performance of UASB reactor
The reactor was started up with a HRT of 12 h for 9 days (Table 1). The removal of sulfide, nitrate, andp-cresol was maintained at 99%, 98%, and 97%,respectively (Stage I in Figure 1). Negligible quantity of nitrite was accumulated in the effluent. Most removed sulfides were converted to elemental sulfur, and the elemental sulfur accumulation rate reached as high as 70%. This result is different from that of the simultaneous removal of sulfide, nitrate and phenol. Beristain-Cardoso,et al.[10]reported that the simultaneous removal of sulfide,nitrate and phenol could be achieved in one reactor and the sulfate was the main end product of sulfide oxidation.From the 10thday on, the concentration of all compounds in the influent was doubled and maintained for another 9 days. The efficiency for removal of sulfide, nitrate andp-cresol was maintained at greater than 98% (stage II in Figure 1), with most end product resulted from sulfide oxidation being elemental sulfur (about 70%).
From the 20thday on, the HRT of the UASB rector was reduced from 12 h to 10 h to increase the loading rates to 240 gS/(m3·d), 240 gN/(m3·d), and 420 gC/(m3·d),respectively. The efficiency for removal of sulfide, nitrate andp-cresol was maintained at 99%, 98% and 92%,respectively. The elemental sulfur accumulation was maintained at 70%. At the same time little nitrite was observed in the effluent.
On day 30, the HRT was further reduced to 7h to increase the loading rates to 340 gS/(m3·d), 340 gN/(m3·d), and 600 gC/(m3·d), respectively. The efficiency for removal of nitrate was maintained at 95% while that of sulfide and p-cresol reduced to 80% and 60%, respectively. The elemental sulfur accumulation rate decreased to 20% at this stage. The UASB reactor performance was seriously deteriorated under these loadings.
3.2 Effects of C/S ratio
With C/S being equal to 0.75:1, the efficiency for removal of sulfide, nitrate andp-cresol was all in excess of 96%. Little nitrite was accumulated in the reactor.Most sulfides removed thereby were converted to sulfate and the elemental sulfur accumulation rate was only about 10% (Figure 2b). When the C/S ratio was equal to 1.0:1, the efficiency for removal of sulfide, nitrate andp-cresol was all maintained at greater than 96%, but the elemental sulfur accumulation was increased to 23%.When the C/S ratio was equal to 1.5:1, the elemental sulfur accumulation rate further increased to 50%. Upon further increasing the C/S ratio to 1.75:1, the elemental sulfur accumulation rate reached as high as 70%, while the efficiency for removal of sulfide, nitrate andp-cresol was all maintained at >96%. This phenomenon indicated that the C/S ratio could affect the end product of sulfide oxidation. Higher C/S ratio resulted in higher elemental sulfur accumulation.
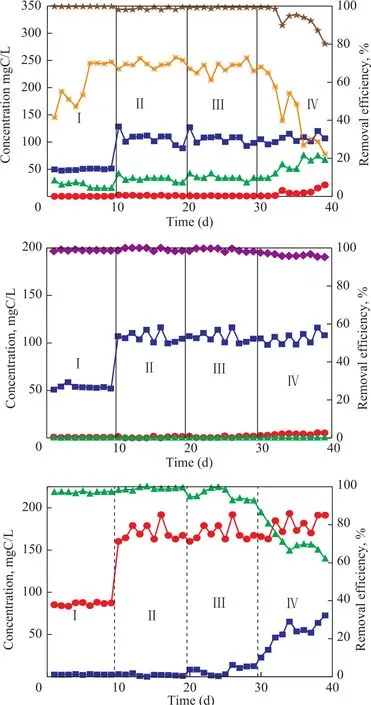
Figure 1 Performance of UASB reactor for simultaneous removal of S2−(a), NO3−-N (b), and p-phenol-C (c)
3.3 Microbial community
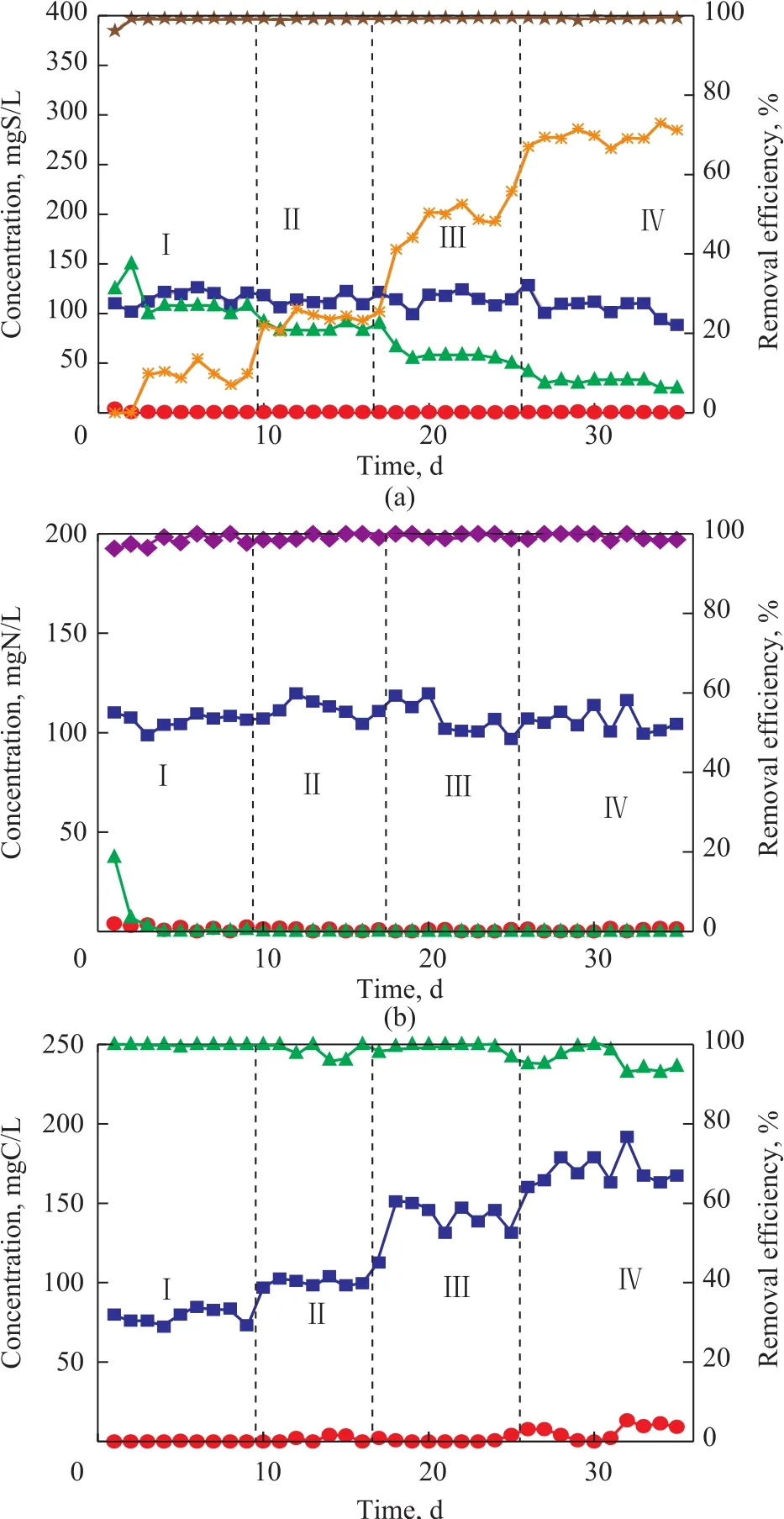
Figure 2 Effects of C/S ratio on UASB performance for simultaneous removal of S2− (a), NO3−-N (b), and p-cresol-C (c)
The parameters related to the alpha diversity of microbial community for each sample at a distance cutoff level of 0.03 are shown in Table 2. The phylogenetic classification of bacterial 16S rRNA sequence from the sludge samples at different loadings was illustrated based on the phylum shown in Figure 3 and on the genus depicted in Table 3.The bacterial sequences affiliated withProteobacteria,Firmicutes,Bacteroideteswere abundant in all samples,and followed a decreasing order:Proteobacteria>Firmicutes>Bacteroidetes. When the influent loading increased from 100 gS/(m3·d), 100 gN/(m3·d) and 150 gC/(m3·d) (stage I in Figure 1) to 240 gS/(m3·d),240 gN/(m3·d) and 420 gC/(m3·d) (stage III in Figure 1),the abundance ofProteobacteriadecreased from 62.95%to 49.40%. On the contrary, the abundance ofFirmicutesandBacteroidetesincreased from 14.97% and 11.25% to 30.39% and13.55%, respectively. When the loading rate was further increased to 340 gS/(m3·d), 340 gN/(m3·d)and 600 gC/(m3·d) (stage IV), the abundance ofProteobacteriafurther decreased to 34.46% and that ofFirmicutesfurther increased to 53.31%. However the abundance ofBacteroidetesdecreased to 6.57%.
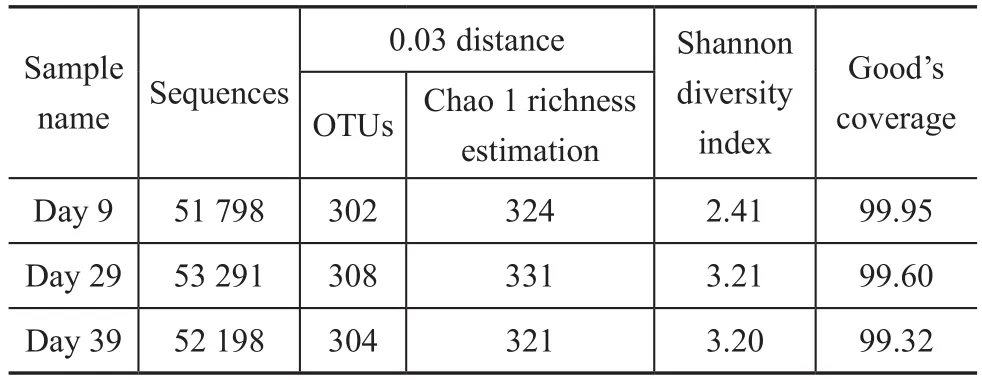
Table 2 Richness and diversity of the three samples based on 0.03 distance
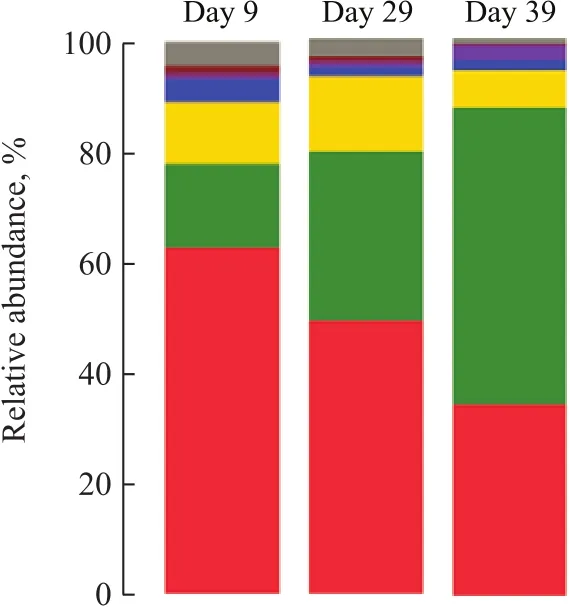
Figure 3 Taxonomic classification of bacterial communities at phylum level at sul fi de loading of 100 gS/(m3·d), 240 gS/(m3·d), and 340 gS/(m3·d). Phylum making up less than 0.5%of total composition in the sample was classified as “others”—Proteobacteria; —Chloroflexi; —Chlorobi; —Firmicutes;—Actinobacteria; —Others; —Bacteroidetes; —Synergistetes
Phylum sequences affiliated withProteiniclasticum,Pseudomonas,Rhizobium,Simplicispira,Arcobacter,Acetoanaerobium,Fusibacter,Petrimonas,Family_XI_unclassified,Sulfurimonas,Aeromonas,Sedimentibacter,Chryseobacterium,Brucella,Arenimonas,Sulfurovum,Paracocccus,SulfurospirillumandThauerawere the main genera detected. The generaPseudomonas,SimplicispiraandRhizobiumwere reported commonly to be hetetrophic denitrifiers[11-13]. The genusPseudomonasaccounted for 25.0% at a loading rate (stage I) of 100 gS/(m3·d), 100 gN/(m3·d), and 150 gC/(m3·d), and decreased to 11.11% and 8.63%, respectively, at stage III and stage IV. A similar pattern was noted for the genusSimplicispira, which was 5.34% at stage I and increased to 7.78% at stage III, then decreased to 0.01% at stage IV.On the contrary, the abundance ofRhizobiumwas 0.91%at stage I and increased to 0.98% and 17.7% at stage III and stage IV, respectively.
The OTUs most closely related toArcobacter,Sulfurimonas,SulfurovumandSulfurospirillum, and the common autotrophic denitrifiers in DSR sludge[14-16], were 0.01%,10.52%, 3.84% and 3.01%, respectively, in the stage I and decreased to 11.63%, 0.41%, 0.09% and 0.53%,respectively, in stage III. Upon further increasing the influent loading rate to 340 gS/(m3·d), 340 gN/(m3·d),and 600 gC/(m3·d), the abundance ofArcobacter,Sulfurimonas,SulfurovumandSulfurospirillumfurther decreased to <0.02%. The sharp decrease of autotrophic denitrifiers might result in the deterioration of the tested UASB reactor.
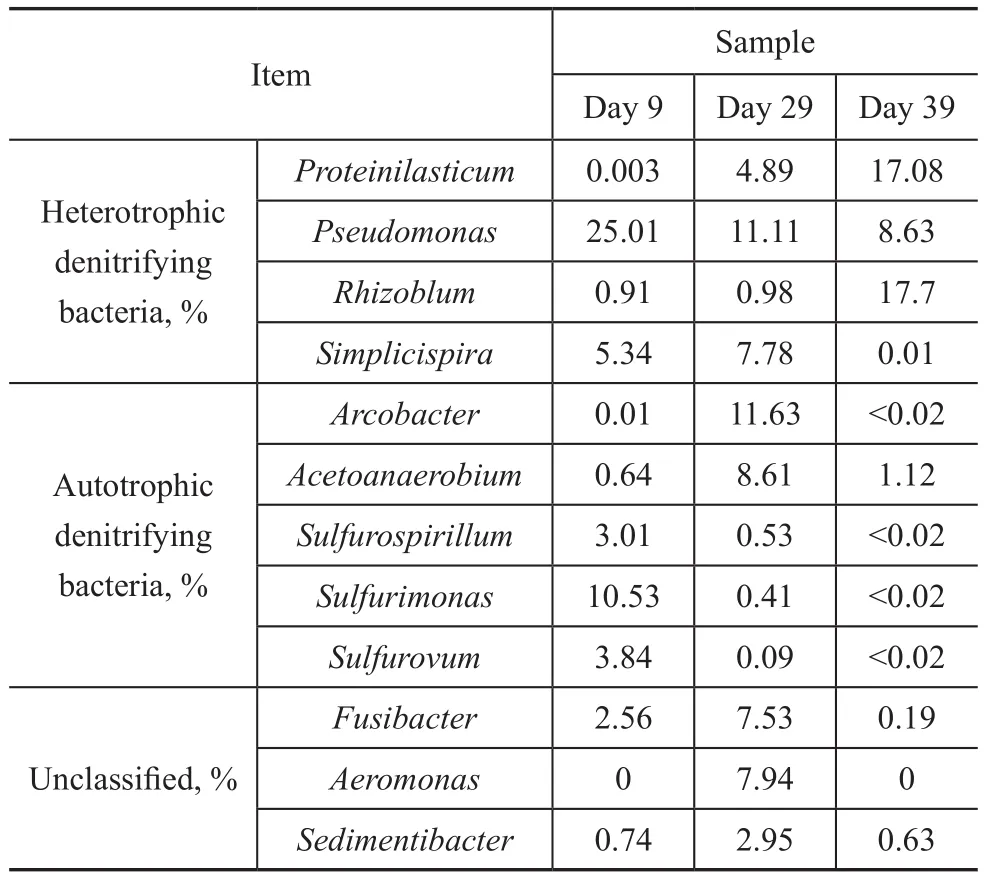
Table 3 Taxonomic classification of bacterial communities at genus level at a sulfide loading of 100 gS/(m3·d), 240 gS/(m3·d), and 340 gS/(m3·d)
4 Conclusions
Simultaneous removal of sulfide, nitrate andp-cresol was achieved in an UASB reactor at loading rates of 340 gS/(m3·d), 340 gN/(m3·d) and 600 gC/(m3·d),respectively. Most of removed sulfide was converted to elemental sulfur, which was different from that of phenol.The ratio of C/S could affect the S0accumulation rate and higher C/S ratio is favorable to the S0accumulation.StrainsPseudomonassp.,Simplicispirasp., andRhizobiumsp. could likely yield heterotrophs. StrainsArcobactersp.,Sulfurimonassp.,Sulfurovumsp. andSulfurospirillumsp. were the autotrophs required for the proposed UASB system. The faster decline of autotrophic denitrifiers (Arcobactersp.,Sulfurimonassp.,Sulfurovumsp.,Sulfurospirillumsp) may lead to the deterioration of the tested UASB reactor.
Acknowledgements: The research was supported by the National Natural Science Foundation of China under Grant No.21307160 and the Fundamental Research Funds for the Central Universities under Grant No. 16CX02040A.
[1]Flyvbjerg J, Jørgensen C, Arvin E, et al. Biodegradation of ortho-cresol by a mixed culture of nitrate-reducing bacteria growing on toluene[J]. Applied and Environmental Microbiology, 1993, 59(7): 2286–2292
[2]Agency for Toxic Substances and Disease Registry(ATSDR). Toxicological pro file for cresols. Atlanta: Public Health Service, U.S. Department of Health and Human Services, 1990
[3]Basheer F, Farooqi I H. Biodegradation ofp-cresol by aerobic granules in sequencing batch reactor[J]. Journal of Environmental Sciences, 2012, 24: 2012–2018
[4]OldhamW K, Rabinowitz B. Development of biological nutrient removal technology in western Canada[J]. Journal of Environmental Engineering and Science, 2002, 1(1): 33–43
[5]Beristain-Cardoso R, Texier AC, Alpuche-Solis A, et al.Phenol and sulfide oxidation in a denitrifying biofilm reactor and its microbial community analysis[J]. Process Biochem, 2009, 44(1): 23–28
[6]Liu C, Han K, Lee D J, et al. Simultaneous biological removal of phenol, sulfide, and nitrate using expanded granular sludge bed reactor[J]. Applied and Environmental Microbiology, 2015, 100(9): 4211–4217
[7]Francisco J C, Meza-Escalante ER, Texier AC, et al.Kinetic limitations during the simultaneous removal of p-cresol and sulfide in a denitrifying process[J]. Journal of Industrial Microbiology & Biotechnology, 2009, 36(11):1417–1424
[8]Liu C, Zhao C, Wang A, et al. Denitrifying sulfide removal process on high-salinity wastewaters[J]. Appl Microbiol Biotechnol, 2015, 99(15): 6463–6469
[9]APHA Standard Methods for the Examination of Water and Wastewater[S]. 21st edn., American Public Health Association, Washington, 2005
[10]Beristain-Cardoso R, Texier A C, Sierra-Alvarea R, et al.Effect of initial sulfide concentration on sulfide and phenol oxidation under denitrifying conditions[J]. Chemosphere,2009, 74: 200–205
[11]Liu B, Mao Y, Bergaust L, et al. Strains in the genus Thauera exhibit remarkably different denitrification regulatory phenotypes[J]. Environ Microbiol, 2013,15(10): 2816–2828
[12]Feng L, Chen K, Han D, et al. Comparison of nitrogen removal and microbial properties in solid-phase denitrification systems for water purification with various pretreated lignocellulosic carriers[J].Bioresour Technol,2017, 224: 236–245
[13]Rungkitwatananukul P, Nomai S, Hirakata Y, et al.Microbial community analysis using MiSeq sequencing in a novel con fi guration fl uidized bed reactor for effective denitri fi cation[J]. Bioresour Technol, 2016, 221: 677–681
[14]Wang A, Du D, Ren N, et al. An innovative process of simultaneous desulfurization and denitrification by Thiobacillus denitrification[J]. Journal of Environmental Science and Health A,2005, 40(5): 1939–1949
[15]Huang C, Li Z, Chen F, et al. Microbial community structure and function in response to the shift of sulfide/nitrate loading ratio during the denitrifying sulfide removal process[J]. Bioresour Technol, 2015, 197(2): 227–234
[16]Lee D J, Wong B T, Adav S S. Azoarcus taiwanensis sp.nov., adenitrifying species isolated from a hot spring[J].Appl Microbiol Biotechnol, 2013, 98(3): 1–7
杂志排行
中国炼油与石油化工的其它文章
- Influence of Initial Water Content on Synthesis of Silicalite-1 Zeolite
- Study on Catalytic Alkylation of Benzene with Methanol over ZSM-22 and ZSM-35
- Preparation of Novel Dechlorination Adsorbent and Study on Its Adsorption Mechanism
- The Function of β Zeolite for Enhancing the Propylene Yield in FCC Process
- Synthesis and Tribological Properties of Graphene-Copper Nanoparticle Composites as Lithium Grease Additive
- Preparation of AgCeY Zeolite Using Microwave Irradiation and Its Adsorptive Desulfurization Performance
