蒙原羚朊蛋白基因的序列分析和多态性研究
2017-09-19王伊琴包勇敢荆文魁陈少博云泽龙秦贞奎杨利峰赵德明
王伊琴 包勇敢 荆文魁 杨 帆 陈少博云泽龙 秦贞奎 杨利峰 赵德明
(1.内蒙古二连浩特出入境检验检疫局,二连浩特,011100;2.中国检验检疫科学研究院,北京,100123;3.中国农业大学动物医学院国家动物海绵状脑病实验室,北京,100094)
蒙原羚朊蛋白基因的序列分析和多态性研究
王伊琴1包勇敢1荆文魁1杨 帆1陈少博1云泽龙1秦贞奎2杨利峰3赵德明3
(1.内蒙古二连浩特出入境检验检疫局,二连浩特,011100;2.中国检验检疫科学研究院,北京,100123;3.中国农业大学动物医学院国家动物海绵状脑病实验室,北京,100094)
稿件运行过程
蒙原羚; 朊蛋白基因; 序列分析; 基因多态性; 朊病毒病; 种间屏障
朊蛋白病(prion protien disease)又称为传染性海绵状脑病(transmissible spongiform encephalopathies,TSE),是由致病性朊蛋白引起的一种致死性神经退行性疾病。可感染多种属动物,其中疯牛病、羊痒病和人的克雅氏病是最常见的海绵状脑病[1]。朊蛋白通过GPI锚定于细胞膜表面的糖蛋白,单拷贝PrP基因编码,整个开放阅读框位于单一外显子内。Prusiner“唯蛋白”理论表明,海绵状脑病的致病因子是一种编码宿主蛋白的PrPC转变为异常的具有致病性的PrPSc,二者都具有相同的氨基酸序列,只是空间结构发生变化,由正常的以α螺旋为主的结构(PrPC)转变为以β折叠为主的结构(PrPSc),在脑部沉积而产生致病性[2-3]。该病可感染多种属动物,其中以反刍动物最为易感,还可以感染人,主要引起人的库鲁病(Kuru),克-雅氏病(creutzfeldt-Jakob disease,CJD)及致死性家族失眠症(fatal familial insomnia,FFI)等。近年来发现的一种人新型克-雅氏病(variant Creutzfeldt-Jakob disease,vCJD)与原来的CJD 疾病在潜伏期、发病人群、病理变化等方面有明显差异,目前科学家们根据流行病学调查推测该病的发生可能源于动物的TSE疾病,食用了患有牛海绵状脑病(Bovine spongiform encephalopathies,BSE)病牛的牛肉,是该病发生的主要原因[2]。
虽然朊蛋白的致病机理尚不清楚,但朊蛋白病的潜伏期、遗传易感性及种间屏障等都与朊蛋白基因氨基酸的多态性相关[4-7 ]。朊病常常是自发的异常性感染或散发性发生。现已证实,除了羊的痒病、牛的疯牛病和人的克雅氏病易感外,大型猫科动物和外来的有蹄动物也易感。对人类健康的危害和疾病控制也是近年来研究的重要课题。PrP的开放阅读框内的多态性与TSE的易感性和致病性有关,如人的克雅氏病、羊痒病[8-9]。
蒙原羚(Procapragutturosa)分布于蒙古、俄罗斯和中国。其性喜群栖,一般随着牧草的生长情况而游动。由于蒙古高原的牛、羊大多是散养方式,这样牛、羊、蒙原羚之间接触机会增多。此外,由于蒙原羚肉可食用,蒙原羚角可入药,感染TSE的蒙原羚产品极易流入食物链,有感染人和动物的潜在风险。因此,关于蒙原羚的TSE的研究变得异常重要。
目前,关于蒙原羚的朊蛋白基因序列的分析和多态性的研究均尚未见报道,本研究通过对26只蒙原羚朊蛋白基因进行克隆与序列测定,由795/771 bp 碱基编码了264/256个氨基酸的前体蛋白,包括N端的24个氨基酸信号肽和C端的22个氨基酸的GPI信号肽[3,10-11],并对其进行基因多态性分析和TSE易感性预测,为TSE的发病机理的相关研究奠定基础。
1 材料与方法
1.1材料
1.1.1试剂
基因组DNA提取试剂盒购于BioDev-Tech公司,DNA凝胶回收试剂盒及pGEM-T Easy 载体购于Promega公司;Taqplus DNA聚合酶、限制性内切酶EcoR I及DNA Marker为TaKaRa公司产品;大肠杆菌感受态细胞购于TransGene Biotech公司,其他试剂均为国产分析纯。
1.1.2样品
本试验是在无菌条件下采取了26份健康蒙原羚的EDTA抗凝血血样。试验研究的蒙原羚均健康,约2~3岁,来自内蒙古自治区呼和浩特市大青山野生动物园。
1.2方法
1.2.1引物
根据GenBank中已发表序列(EU 224471),应用Primer premier 5.0设计特异性引物,由上海生工合成。
上游引物:5′-ATGGTGAAAAGCCACATAGGCAGTTG-3′;
下游引物:5′5′-CTATCCTACTATGAGAAAAATGAGGAAAG-3′。
1.2.2 Prnp的扩增
该引物扩增了绵羊朊蛋白开放阅读框ORF由795/771 bp核苷酸,共编码了264/256个氨基酸。基因组总DNA的提取按试剂盒方法进行。PCR 反应在25 μL体系中进行,其中含有:基因组DNA 200 ng;引物 0.5 μM;dNTP 200 μM;Taqplus DNA Polymerase(5 U/μL)0.5 μL;0.1 M 10×Ammonium Buffer;去离子水17 μL,进行PCR扩增,循环条件如下:94℃预变性5 min、94℃变性30 s、61℃退火40 s、72℃延伸40 s、共循环36 次、最后于72℃延伸8 min。
1.2.3 PCR产物的克隆
扩增产物纯化按试剂盒纯化,纯化后PCR扩增产物与pGEM-T easy vector进行连接,4℃水浴过夜。将上述连接产物转化大肠杆菌DH5α,涂布于含有氨苄青霉素,X-gal和IPTG的LB琼脂平板,于37℃温箱中倒置培养20 h,随机挑取白色菌落进行筛选。挑取单个菌落接种于含氨苄青霉素的LB 培养液中,37℃振荡培养过夜。利用质粒提取试剂盒制备质粒,然后对其进行酶切和PCR鉴定。
1.2.4序列测定及分析
纯化后PCR扩增产物直接测序或克隆至pGEM-T easy vector后,每个样品挑取3~4个克隆进行测序。并利用DNAMAN软件(Version 5.2.2)进行序列比较和同源性分析。
2 结果
2.1 PCR扩增、克隆和酶切鉴定
PCR扩增产物经1%琼脂糖凝胶电泳分析,条带大小与扩增片段基本一致约为795/771 bp(图1),初步证实为蒙原羚Prnp基因。PCR产物纯化后克隆到载体中,转化后挑取阳性菌落,经EcoRⅠ酶切可获得3 000和795或771 bp左右2个条带(Pgem-T Easy载体两端分别存在1个EcoRⅠ酶切位点),这与载体和插入目的的片段大小正好相符(图2);同时应用上、下游引物在重组质粒中PCR扩增出大小约为795或771 bp的条带,结果表明扩增片段已经插入载体中(图略)。
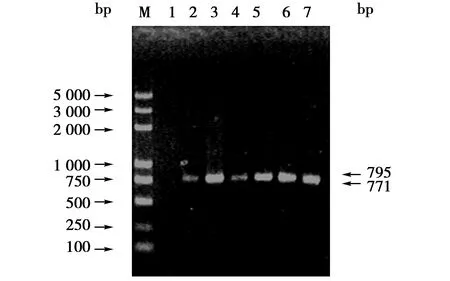
图1 Prnp的PCR产物琼脂糖凝胶电泳结果Fig.1 Agarose gel electrophorsis result of Prnp PCR products 注:M:DL 2 000 DNA 相对分子质量标准,2~7:不同样品PCR产物 Note:M:DL 2 000 plus DNA marker,2-7:PCR products from different samples
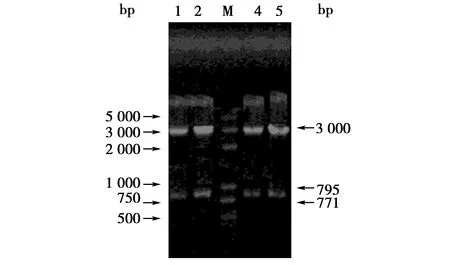
图2 阳性克隆质粒的酶切鉴定Fig.2 Analysis of plasmids digested by EcoR I 注:1、2、4、5:不同样品重组质粒EcoR I酶切鉴定结果,M:DL 2 000相对分子质量标准 Note:1,2,4,5:products from different recombinant plasmids digested by EcoR I,M:DL 2 000 plus DNA marker
2.2蒙原羚Prnp基因的序列分析及其多态性
蒙原羚Prnp的ORF 795/771 bp核苷酸编码了264/256氨基酸,蒙原羚朊蛋白序列分析并提交于GenBank(AB473602-AB473615)。应用DNAMAN、DNASTAR进行序列分析比较,26只蒙原羚的朊蛋白基因高度同源。蒙原羚prion protein的开放阅读框内有6个氨基酸的多态性位点,氨基酸置换情况分别为119(N→S),143(S→G),160(Y→H),172(V→A),182(N→S)和221(V→A)(表1)。
蒙原羚与其他11种动物朊蛋白氨基酸的比较(图3)。蒙原羚朊蛋白同源性比较结果分别是汤氏瞪羚(Eudorcasthomsonii,EU032301,100%)、印度羚(Antilopecervicapra,AY720706,100%)、牛(Bostaurus,EU224471,98.5%)、大羚羊(Tragelaphusoryx,EF165082,98.1%)、欧洲狍(Capreoluscapreolus,AY639096,96.3%)、斑纹角马(Connochaetes,EF165086,96.3%)、绵羊(Ovisaries,M31313,95.9%)、山羊(Caprahircus,EU032305,95.9%)、黇鹿(Cervusdama,AY639094,95.6%)、人(Homosapiens,NM183079,86.9%)、小家鼠(Musmusculus,NM011170,84.3%)。蒙原羚朊蛋白基因有缺失的基因型与斑纹角马、欧洲狍、绵羊、山羊高度同源,分别为99.3%、99.3%、98.9%、98.9%(图3,图4)和13种动物朊蛋白同源性比较的进化树见图5。
表1蒙原羚朊蛋白基因多态性位点
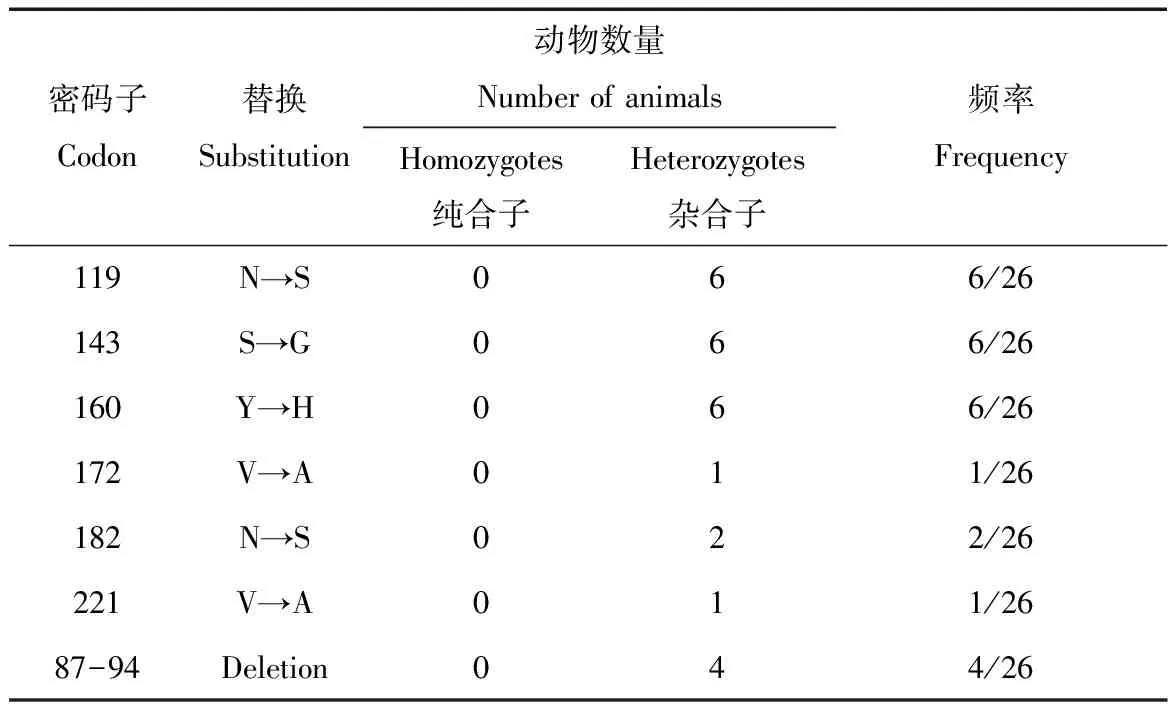
Tab.1 Frequencies of Mongolian gazelle Prnp polymorphism
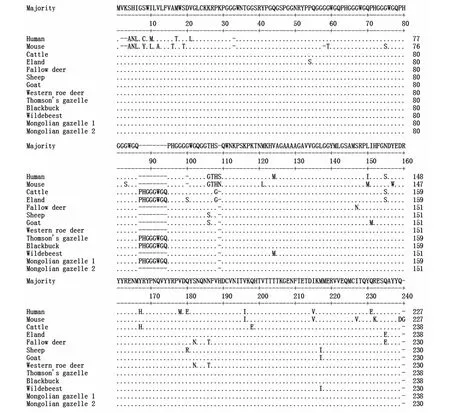

图3 13种哺乳动物朊蛋白氨基酸的比较Fig.3 Amino acid alignment of thirteen mammalian prion proteins 注:人(Human,Homo sapiens,NM183079),小家鼠(Mouse,Mus musculus,NM011170),牛(Cattle,Bos taurus,EU224471),大羚羊(Eland,Tragelaphus oryx,EF165082),黇鹿(Fallow deer,Cervus dama,AY639094),绵羊(Sheep,Ovis aries,M31313),山羊(Goat,Capra hircus,EU032305),欧洲狍(Western roe deer,Capreolus capreolus,AY639096),汤氏瞪羚(Thomson’s gazelle,Eudorcas thomsonii,EU032301),印度羚(Blackbuck,Antilope cervicapra,AY720706),和斑纹角马(Wildebeest,Connochaetes taurinus,EF165086)与蒙原羚(Mongolian gazelles,Procapra gutturosa,AB473611 和 AB473604)的氨基酸的比较。用破折号表示序列的缺失、用点表示氨基酸序列的一致性Note:The deduced amino acid sequence of Human(Homo sapiens,NM183079),Mouse(Mus musculus,NM011170),Cattle(Bos taurus,EU224471),Eland(Tragelaphus oryx,EF165082),Fallow deer(Cervus dama,AY639094),Sheep(Ovis aries,M31313),Goat(Capra hircus,EU032305),Western roe deer(Capreolus capreolus,AY639096),Thomson’s gazelle(Eudorcas thomsonii,EU032301),Blackbuck(Antilope cervicapra,AY720706),and Wildebeest(Connochaetes taurinus,EF165086)were compared with Mongolian gazelles(Procapra gutturosa,AB473611 and AB473604).Deletions are indicated by dashes and sites identical to the consensus sequence are denoted by dots
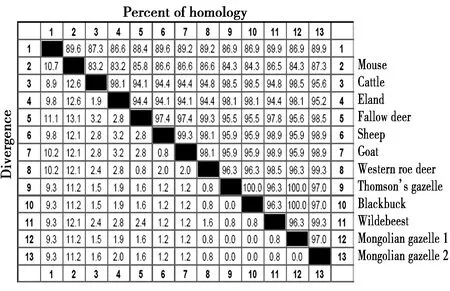
图4 13种哺乳动物朊蛋白的同源性的比较Fig.4 Sequence homology of thirteen mammalian prion proteins 注:1.人(NM183079),2.小家鼠(NM011170),3.牛(EU224471),4.大羚羊(EF165082),5.黇鹿(AY639094),6.绵羊(M31313),7.山羊(EU032305),8.欧洲狍(AY639096),9.汤氏瞪羚(EU 032301),10.印度羚(AY720706),11.斑纹角马(EF165086),12.蒙原羚(AB473611和AB473604) Note:1.Human(NM183079),2.Mouse(NM011170),3.Cattle(EU224471),4.Eland(EF165082),5.Fallow deer(AY639094),6.Sheep(M31313),7.Goat(EU032305),8.Western roe deer(AY639096),9.Thomson’s gazelle(EU032301),10.Blackbuck(AY720706),11.Wildebeest(EF165086),12.Mongolian gazelle(AB473611),13.Mongolian gazelle(AB473604)
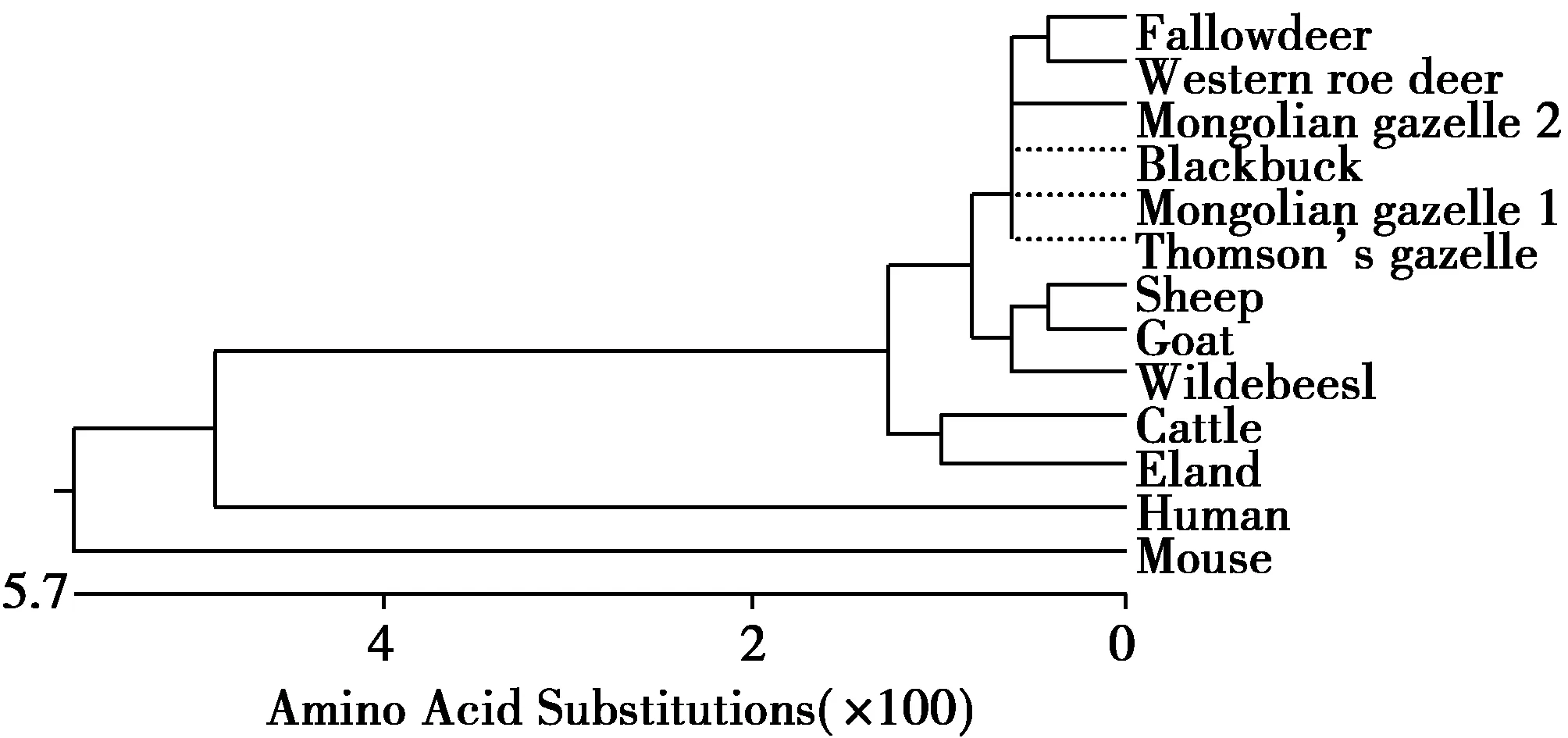
图5 13种哺乳动物朊蛋白同源性比较的进化树Fig.5 Phylogenetic tree of thirteen mammalian prion proteins 注:Human(NM183079),Mouse(NM011170),Cattle(EU224471),Eland(EF165082),Fallow deer(AY639094),Sheep(M31313),Goat(EU032305),Western roe deer(AY639096),Thomson’s gazelle(EU 032301),Blackbuck(AY720706),Wildebeest(EF165086),Mongolian gazelle1(AB473611),Mongolian gazelle 2(AB473604)
朊蛋白基因的N端有几个数目不等的八肽重复区,大多数哺乳动物中有5个八肽重复区,而牛是有6个。在本研究中,26只蒙原羚中大多数(84.6%)在朊蛋白基因的N端有6∶6重合子基因型,少数(15.4%)是6∶5杂合子基因型,未见有5∶5的基因型。具体缺失的位置在R5(87-94),缺失1个八肽重复区(表2)。
表2 蒙原羚的八肽重复区
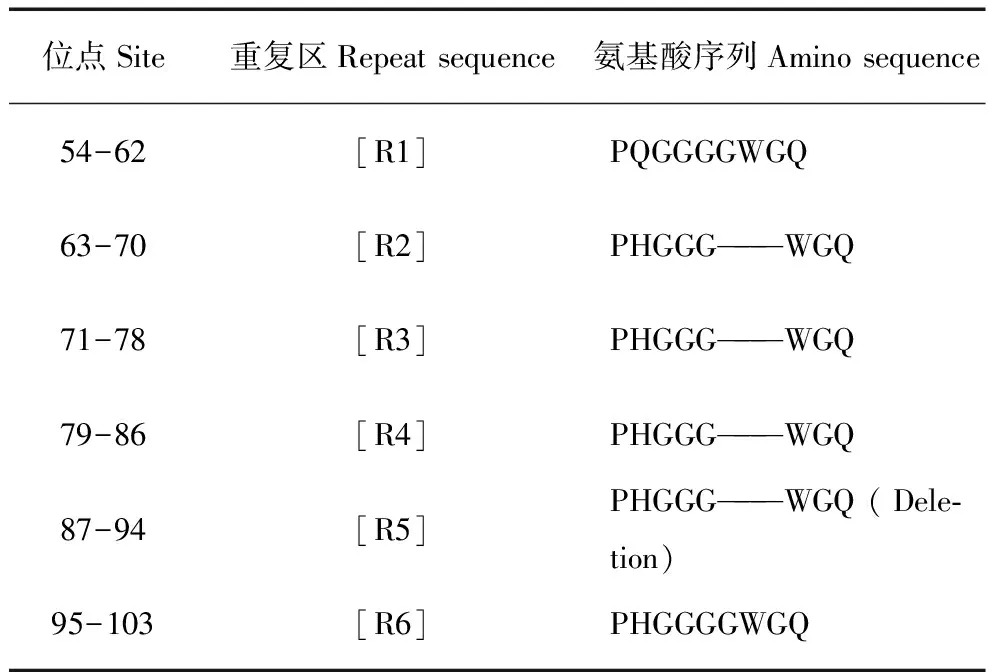
Tab.2 Octapeptide repeats in gPrnp
注:重复区缺失发生在R5 87-94位的氨基酸
Note:The octapeptide repeats between R4 and R6 was deleted
3 讨论
朊蛋白供体与受体之间的PrP同源性与物种屏障有关。二者的同源性越高,就越易突破种间屏障而易感[12-14]。已经证实了转基因鼠中的187、189、206、208的氨基酸所形成的表位参与了突破种间屏障而感染BSE[15-16]。蒙原羚的朊蛋白基因与绵羊、鹿、牛的基因序列高度同源,分别为95.9%、95.6%和98.5%。蒙原羚在6个位点发生氨基酸的置换分别为119(N→S),143(S→G),160(Y→H),172(V→A),182(N→S)和221(V→A)。这些多态性位点位于PrP的C端球形结构域,影响着人和动物的种间屏障。
各种数据显示Prnp的多态性与朊病的易感性、潜伏期和种间屏障有密切关系[5,17-19]。羊的136、154、171三个位点的氨基酸与羊痒病有关。136(A→V)、154(R→H)、171(Q→R/H/K)3个位点的等位基因编码的氨基酸对羊痒病易感性影响最大。在136、154、171的3个位点编码的氨基酸分别是Val/Arg/Gln(VRQ),对羊痒病非常易感,其多态性与羊痒病的潜伏期、易感性有关。在136、154、171氨基酸分别是Ala/Arg/Arg(ARR)时,与羊痒病的抗性有关。而在136、154、171氨基酸分别是Ala/Arg/Gln或His(ARQ或ARH)时,对该病有中度易感性(medium-high susceptibility)[20-23]。落基山马鹿(Cervuselaphusnelsoni)和麋鹿(Elaphurusdavidianus)发生的慢性消耗性疾病(CWD),与朊蛋白基因的多态性相关。在132位点是甲硫氨酸时,麋鹿或鹿发生CWD的易感性增加[24]。CJD、GSS、FFI的发生与人的朊蛋白基因的ORF的氨基酸的变化有关。人的朊蛋白基因在129位点的氨基酸是甲硫氨酸的纯合子型时,人对VCJD有易感性[6,25]。研究证实,15例波兰CJD患者,在129位点有73.3%是甲硫氨酸的纯合子型,13.3%Val纯合子型、13.3%Met /Val杂合子型,而119例正常波兰人,在129位点是45%甲硫氨酸的纯合子型[6,25]。研究证明,羊痒病感染神经细胞瘤转染人或鼠的PrP嵌合体基因,在位点167、171、214、和218氨基酸替换时,可以抗PrPSc的形成[26]。因此,人和动物PRP的C-端球形结构域与朊病的易感性相关,但是蒙原羚朊蛋白基因在C-端球形结构域的多态性是否有朊病的易感性或抗性还需进一步探讨。
在本研究中,大部分(85.4%)蒙原羚的八肽重复区是6∶6基因型,而少部分(14.6%)是6∶5基因型(表2)。蒙原羚的八态重复区是介于牛和羊之间,牛大多数是6个重复区,羊大多数是有5个重复区(图3)。某些遗传性朊病毒病,例如遗传性朊病如家族性Creutzfeldt-Jakob病(fCJD)、GSS综合征均与Prnp八肽重复区插入突变相关,插入突变超过8个以上增加了CJD易感性[27],在朊病病例中,朊蛋白基因八肽重复数目越多发病越早,病程越短[28-29]。对于动物来说,八肽插入突变数目的多少与动物朊病的易感性尚无定论[27]。目前,蒙原羚尚未有朊病的报道。
总之,该研究结果显示,蒙原羚朊蛋白基因种间高度同源,蒙原羚在6个位点发生氨基酸的置换分别为119、143、160、172、182和221,通过对13种蒙原羚等哺乳动物的Prnp分析,与汤氏瞪羚、印度羚100%同源,与牛、大角斑羚同源性高于98%,此外,C-端球形结构域的R4与R6之间(87-94)缺失1个八肽重复区。分别与牛和羊的同源性相比较,八肽重复区的长度与牛相同,故蒙原羚Prnp和牛的同源性高于和羊的同源性。目前,尚未有蒙原羚感染TSE的报道,对蒙原羚Prnp多态性的研究,可能对TSE的发病机制、朊病的抗性基因和跨种间传播将有重要的意义。
[1] Prusiner S B.Molecular biology and pathogenesis of prion diseases[J].Trends in Biochemical Sciences,1996,21(12):482-487.
[2] Hill A F,Desbruslais M,Joiner S,et al.The same prion strain causes vCJD and BSE[J].Nature,1997,389(6650):448-500,526.
[3] Prusiner S B.Prions[J].Proceedings of the National Academy of Sciences of the United States of America,1998,95(23):13363-13383.
[4] Asante E A,Linehan J M,Desbruslais M,et al.BSE prions propagate as either variant CJD-like or sporadic CJD-like prion strains in transgenic mice expressing human prion protein[J].The EMBO Journal,2002,21(23):6358-6366.
[5] Bossers A,Schreuder B E C,Muileman I H,et al.PrP genotype contributes to determining survival times of sheep with natural scrapie[J].Journal of General Virology,1996,77(10):2669-2673.
[6] Collinge J,Sidle K C L,Meads J,et al.Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD[J].Nature,1996,383(6602):685-690.
[7] Westaway D,Zuliani V,Cooper C M,et al.Homozygosity for prion protein alleles encoding glutamine-171 renders sheep susceptible to natural scrapie[J].Genes & Development,1994,8(8):959-969.
[8] Kirkwood J K,Cunningham A A,Epidemiological observations on spongiform encephalopathies in captive wild animals in the British Isles[J].The Veterinary Record,1994,135(13):296-303.
[9] Lezmi S,Bencsik A,Monks E,et al.First case of feline spongiform encephalopathy in a captive cheetah born in France:PrPscanalysis in various tissues revealed unexpected targeting of kidney and adrenal gland [J].Histochemistry and Cell Biology,2003,119(5):415-422.
[10] Kuroda Y,Maeda Y,Sawa S,et al.Effects of detergents on the secondary structures of prion protein peptides as studied by CD spectroscopy[J].Journal of Peptide Science,2003,9(4):212-220.
[11] Walmsley A R,Hooper N M.Distance of sequons to the C-terminus influences the cellular N-glycosylation of the prion protein.[J].Biochemical Journal,2003,370(1):351-355.
[12] Prusiner S B.Molecular biology of prion diseases[J].Science,1991,252(5012):1515-1522.
[13] Schätzl H M,Da Costa M,Taylor L,et al.Prion protein gene variation among primates[J].Journal of Molecular Biology,1995,245(4):362-374.
[14] Scott M,Foster D,Mirenda C,et al.Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques[J].Cell,1989,59(5):847-857.
[15] Scott M R,Safar J,Telling G,et al.Identification of a prion protein epitope modulating transmission of bovine spongiform encephalopathy prions to transgenic mice[J].Proceedings of the National Academy of Sciences of the United States of America,1997,94(26):14279-14284.
[16] Wopfner F,Weidenhöfer G,Schneider R,et al.Analysis of 27 mammalian and 9 avian PrPs reveals high conservation of flexible regions of the prion protein[J].Journal of Molecular Biology,1999,289(5):1163-1178.
[17] Baylis M,Goldmann W,Houston F,et al.Scrapie epidemic in a fully PrP-genotyped sheep flock[J].Journal of General Virology,2002,83(11):2907-2914.
[18] Goldmann W,Hunter N,Foster J D,et al.Two alleles of a neural protein gene linked to scrapie in sheep[J].Proceedings of the National Academy of Sciences of the United States of America,1990,87(7):2476-2480.
[19] Hunter N,Goldmann W,Marshall E,et al.Sheep and goats:natural and experimental TSEs and factors influencing incidence of disease[J].Archives of Virology,2000,16(S):181-188.
[20] Belt P B G M,Muileman I H,Schreuder B E C,et al.Identification of five allelic variants of the sheep PrP gene and their association with natural scrapie[J].Journal of General Virology,1995,76(3):509-517.
[21] Houston F,Goldmann W,Chong A,et al.Prion diseases:BSE in sheep bred for resistance to infection[J].Nature,2003,423(6939):498.
[22] Hunter N,Moore L,Hosie B D,et al.Association between natural scrapie and PrP genotype in a flock of Suffolk sheep in Scotland[J].The Veterinary Record,1997,140(3):59-63.
[23] McCutcheon S,Hunter N,Houston F.Use of a new immunoassay to measure PrPSclevels in scrapie-infected sheep brains reveals PrP genotype-specific differences[J].Journal of Immunological Methods,2005,298(1/2):119-128.
[24] O’Rourke K I,Besser T E,Miller M W,et al.PrP genotypes of captive and free-ranging Rocky Mountain elk(Cervuselaphusnelsoni)with chronic wasting disease[J].Journal of General Virology,1999,80(10):2765-2769.
[25] Bratosiewicz J,Liberski P P,Kulczycki J,et al.Codon 129 polymorphism of the PRNP gene in normal Polish population and in Creutzfeldt-Jakob disease,and the search for new mutations in PRNP gene[J].Acta Neurobiologiae Experimentalis,2001,61(3):151-156.
[26] Kaneko K,Zulianello L,Scott M,et al.Evidence for protein X binding to a discontinuous epitope on the cellular prion protein during scrapie prion propagation[J].Proceedings of the National Academy of Sciences of the United States of America,1997,94(19):10069-10074.
[27] Goldmann W.PrP genetics in ruminant transmissible spongiform encephalopathies[J].Veterinary Research,2008,39(4):1-14.
[28] Goldfarb L G,Brown P,McCombie W R,et al.Transmissible familial Creutzfeldt-Jakob disease associated with five,seven,and eight extra octapeptide coding repeats in the PRNP gene[J].Proceedings of the National Academy of Sciences of the United States of America,1991,88(23):10926-10930.
[29] Mead S.Prion disease genetics[J].European Journal of Human Genetics,2006,14(3):273-281.
Mongolian gazelle(Procapragutturosa); Prnp; Sequence analysis; Gene polymorphism; Prion disease; Species barrier
传染性海绵状脑病(TSE)是发生于人和动物神经系统的退行性疾病,影响着人类的健康及动物福利。朊蛋白基因的ORF氨基酸的多态性(Prnp)与TSE的潜伏期、遗传易感性及种间屏障有关。在本研究中,首次对蒙原羚(Procapragutturosa)朊蛋白基因的ORF进行全序列分析和多态性研究。研究结果发现,26只蒙原羚Prnp高度同源,与汤氏瞪羚(Eudorcasthomsonii)、印度羚(Antilopecervicapra)、牛(Bostaurus)的SNP的同源性较近,分别为100%、100%和98.5%,然而,斑纹角马(Connochaetestaurinus)、欧洲狍(Capreoluscapreolus)和绵羊(Ovisaries)与八肽重复区的缺失相关,蒙原羚与它们的同源性分别为99.3%、99.3%和98.9% 。蒙原羚在6个位点发生氨基酸的置换分别为119(N→S),143(S→G),160(Y→H),172(V→A),182(N→S)和221(V→A)。在N端的R5(87-94)缺失1个八肽重复区。蒙原羚Prnp序列的特殊性可能影响分子间的相互作用和TSE种间屏障起关键的作用。
Sequences Analysis and Polymorphism Research of the Prion Protein Gene in Mongolian Gazelles (Procapra gutturosa)
Wang Yiqin1Bao Yonggan1Jing Wenkui1Yang Fan1Chen Shaobo1Yun Zelong1Qin Zhenkui2Yang Lifeng3Zhao Deming3
(1.Erlianhot Entry-Exit Inspection and Quarantine,Erlianhot,011100,China;2.Chinese Academy of Inspection and Quarantine,Beijing,100123,China;3.National Animal Transmissible Spongiform Encephalopathies Laboratory,College of Veterinary Medicine,China Agricultural University,Beijing,100094,China)
Prion diseases are a group of human and animal neurodegenerative conditions that are caused by the deposition of an abnormal isoform prion protein (PrPSc)encoded by a single copy prion protein gene (Prnp).In sheep,genetic variations of Prnp were found to be associated with the incubation period,susceptibility,and species barrier to the scrapie disease.We investigated the sequence and polymorphisms of the prion protein gene of Mongolian gazelles (gPrnp).gPrnp gene sequence analysis of blood samples from 26 Mongolian gazelles showed high identity within species.The gPrnp gene was closely related to the Prnp genes of Thomson’s gazelle,blackbuck,and cattle with 100,100,and 98.5% identity,respectively.The gPrnp gene with a deletion was closely related to the Prnp genes of wildebeest,Western roe deer,and sheep with 99.3,99.3,and 98.9% identity,respectively.Polymorphisms of the open reading frame of Prnp as amino acid substitutions were detected at codons 119(N→S),143(S→G)or 160(Y→H),172(V→A),182(N →S)and 221(V→A).There was also deletion of one octapeptide repeat at the N-terminal octapeptide repeat region.The polymorphisms of gPrnp will assist the study of prion disease pathogenesis,resistance,and cross species transmission.
王伊琴,女,49岁,博士,高级兽医师;研究方向,人畜共患病致病机理,主要从事出入境动物检验检疫工作。 E-mail:bygwyq@163.com
2016-10-20
S852.65+9.7
A
修回日期:2016-11-30
发表日期:2017-05-10
2310-1490(2017)02-168-07
