Crystal Structures and DNA Interaction Properties of Niand Ni(Ⅱ) and Cd(Ⅱ)Complexes with a Semicarbazone Ligand Bearing Pyrazine Unit
2017-07-05MAOPanDongZHAOXiaoLeiSHAOZhiPengLIMinWUWeiNaWANGYuan
MAO Pan-DongZHAO Xiao-Lei*,SHAO Zhi-PengLI MinWU Wei-Na*,WANG Yuan
(1College of Chemistry and Chemical Engineering,Henan Polytechnic University,Jiaozuo,Henan 454000,China)
(2School of Materials Science and Engineering,Henan Polytechnic University,Jiaozuo,Henan 454000,China)
Crystal Structures and DNA Interaction Properties of Niand Ni(Ⅱ) and Cd(Ⅱ)Complexes with a Semicarbazone Ligand Bearing Pyrazine Unit
MAO Pan-Dong1ZHAO Xiao-Lei*,1SHAO Zhi-Peng2LI Min2WU Wei-Na*,1WANG Yuan1
(1College of Chemistry and Chemical Engineering,Henan Polytechnic University,Jiaozuo,Henan 454000,China)
(2School of Materials Science and Engineering,Henan Polytechnic University,Jiaozuo,Henan 454000,China)
Two complexes,namely[NiL2]·CH3OH·0.5H2O(1)and[Cd(HL)Cl2](2)(HL=1-(3-methylpyrazin-2-yl) ethylidene-4-phenylsemicarbazide)have been synthesized and characterized by single crystal X-ray diffraction, elemental analysis and IR spectroscopy.X-ray diffraction analysis results show that the central Niion in complex 1 is surrounded by two independent anionic ligands with N2O donor set,thus possesses a distorted octahedral coordination geometry.However,in complex 2,the Cdion with a distorted square pyramidal is coordinated with one neutral tridentate ligand HL and two chloride anions.In addition,the fluorescence spectra indicate that the interactions of the complexes with DNA are stronger than that of the ligand HL.CCDC 1479242:1;1479243:2.
semicarbazone;Nicomplex;Cdcomplex;pyrazine;crystal structure;DNA interaction
As an important class of ligands,Schiff bases play a crucial role in coordination chemistry and have been widely applied in different fields due to their broad spectrum of biological applications[1-13].In recent years,the transition metal complexes of Schiff base derivatives bearing a heterocyclic ring(such as pyrrole[2],pyridine[3-6],pyrazine[7-10]and quinoline[11-13]), involving acylhydrazones[3-6],thiosemicarbazones[9-13]and semicarbazones[7-8,11]have been proved to possess high biological and pharmaceutical activities.Our previousworkalsoshowsthattwobinuclear complexes[Cu2(L)2Cl2]and[Cu2(L)2(OAc)2]with 1-(3-methylpyrazin-2-yl)ethylidene-4-phenylsemicarbazide (HL,Scheme 1)can bind to DNA and have potential pharmaceutical activity[7].
On the other hand,the biological activities of the complexes depend mainly on the metal centers[9-10].It should be noted that metal-ligand synergism could occur in most cases.In addition,several Niand Cdcomplexes should be investigated because both Niand Cdions are closely related to biochemistry, clinicaldiagnostics,as well as environmental pollution[8]. Therefore,as the continuation of our work,Niand Cdcomplexes with HL have been synthesized and structurallydeterminedbysingle-crystalX-ray diffraction.In addition,the interactions between the compounds and ct-DNA have been studied by ethidium bromide(EB)fluorescence probe.
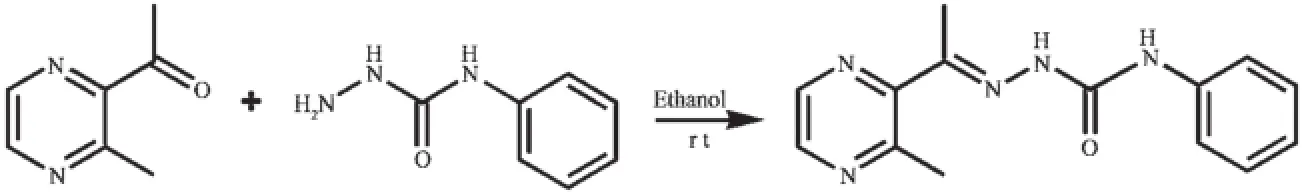
Scheme 1Synthesis route of HL
1 Experimental
1.1 Materials and measurements
Solvents and starting materials for synthesis were purchasedcommerciallyandusedasreceived. Elemental analysis was carried out on an Elemental Vario EL analyzer.PXRD data were recorded on a Philips X′Pert-MPD instrument with Cu Kα radiation (λ=0.154 056 nm)at 293 K.The IR spectra(ν=4 000~400 cm-1)were determined by the KBr pressed disc method on a Bruker V70 FTIR spectrophotometer. The UV spectra were recorded on a Purkinje General TU-1800spectrophotometer.TGanalysiswas measuredonaMettler-ToledoTGA/SDTA851einstrument.The interactions between the complexes and ct-DNA are measured using literature method[13]via emission spectra on a Varian CARY Eclipse spectrophotometer.
1.2 Preparations of complexes 1 and 2
The ligand HL was produced by condension of 1-(3-methylpyrazin-2-yl)ethanone and 4-phenylsemicarbazidefollowingthereportedprocedure[8].The complexes 1 and 2 were generated by reaction of the ligand HL(5 mmol)with equimolar of Ni(NO3)2and CdCl2in methanol solution(10 mL),respectively. Crystals suitable for X-ray diffraction analysis were obtained by evaporating the corresponding reaction solutions at room temperature.
1:brown blocks.Anal.Calcd.for C29H33N10O3.5Ni (%):C:54.74;H:5.23;N:22.01.Found(%):C: 54.98;H:5.32;N:21.86.FTIR(cm-1):ν(N=C-O) 1 615,ν(C=N-N)1 594,ν(C=N)pyrazine1 548.
2:colorlessneedles.Anal.Calcd.for C14H15CdCl2N5O(%):C:37.15;H:3.34;N:15.47. Found(%):C:37.22;H:3.16;N:15.27.FTIR(cm-1): ν(O=C)1 642,ν(C=N-N)1 589,ν(C=N)pyrazine1 544.
1.3 X-ray crystallography
The X-ray diffraction measurement for complexes 1 and 2 were performed on a Bruker SMART APEXⅡCCD diffractometer equipped with a graphite monochromatized Mo Kα radiation(λ=0.071 073 nm) by using φ-ω scan mode.Semi-empirical absorption correction was applied to the intensity data using the SADABS program[14].The structures were solved by direct methods and refined by full matrix least-square on F2using the SHELXTL-97 program[15].All nonhydrogen atomswererefinedanisotropically.The occupancy value of O4 atom in complex 1 is 0.5,and thus the H atoms of which are not added.All the other H atoms were positioned geometrically and refined using a riding model.Details of the crystal parameters,datacollectionandrefinementsfor complexes 1 and 2 are summarized in Table 1.
CCDC:1479242:1;1479243:2.

Table 1Crystal data and structure refinement for complexes 1 and 2
2 Results and discussion
2.1 Crystal structures description
Selected bond distances and angles,hydrogen bonds information for both complexes are listed in Table 2 and 3,respectively.As shown in Fig1a,the central Niion in complex 1 is surrounded by two independent anionic ligands with N2O donor set,thus possessesadistortedoctahedralcoordination geometry:in fact,the N8-Ni1-N3 angle(178.42(13)°) is very close to the theoretical 180°,but the other two,N1-Ni1-O1 and O2-Ni1-N6 are 154.94(14)°and154.48(13)°,respectively,due to the chelation rings strain.The enolization of C=O bond of the ligand can be confirmed by the bond lengths of C-O being 0.126 6(2)and 0.128 8(1)nm,which are in excellent agreementwithpreviouslyknownsemicarbazone complexes[7-8].The distances of Ni-N/O bonds were in the range of 0.200 9(4)~0.213 3(4)nm,comparable with those in some reported complexes with similar donor set[8].In the solid state,two complex molecules linkwitheachotherintoacentrosymmetric moleculardimer via N-H…O hydrogen bonds.In addition,intermolecularN-H…OandN-H...O hydrogen bonds between the complex and crystal methanol molecules are also present(Fig.1c).
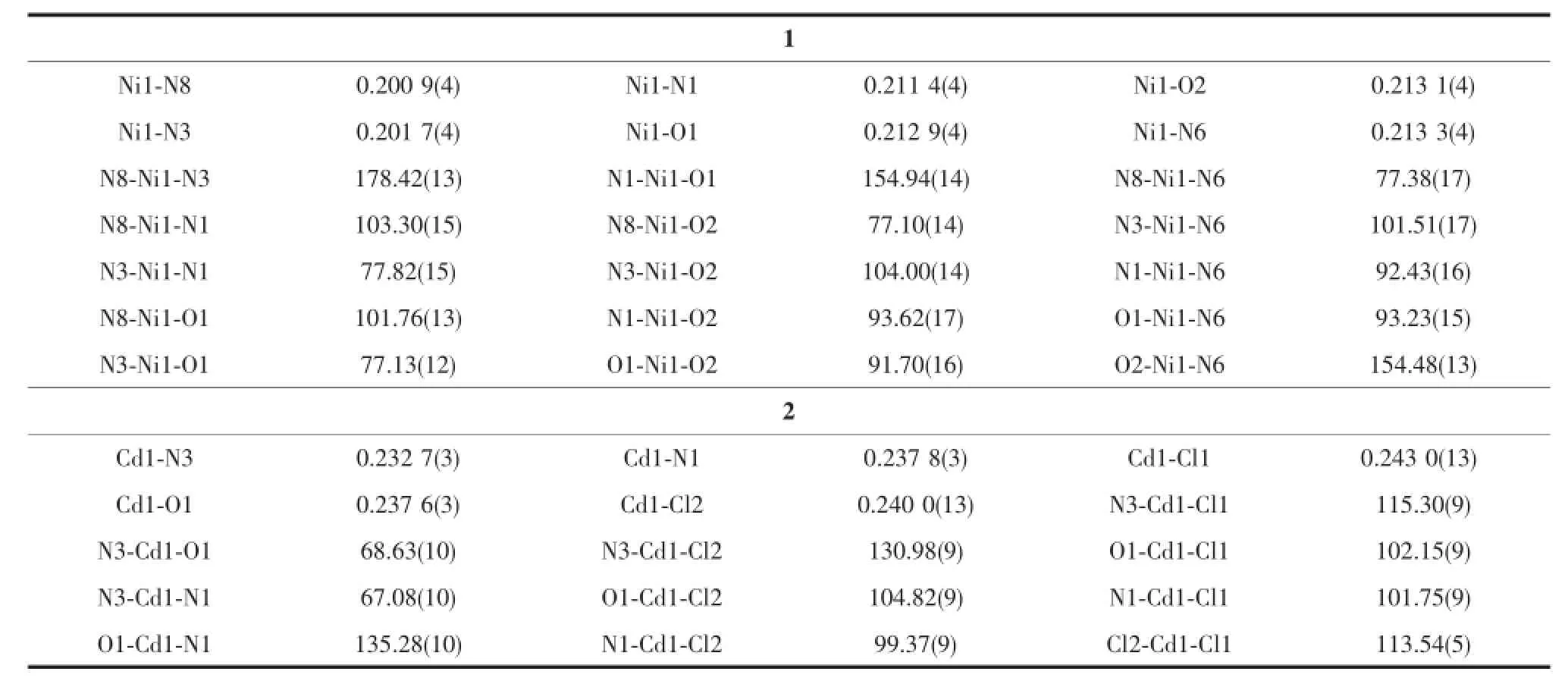
Table 2Selected bond lengths(nm)and angles(°)in complexes 1 and 2
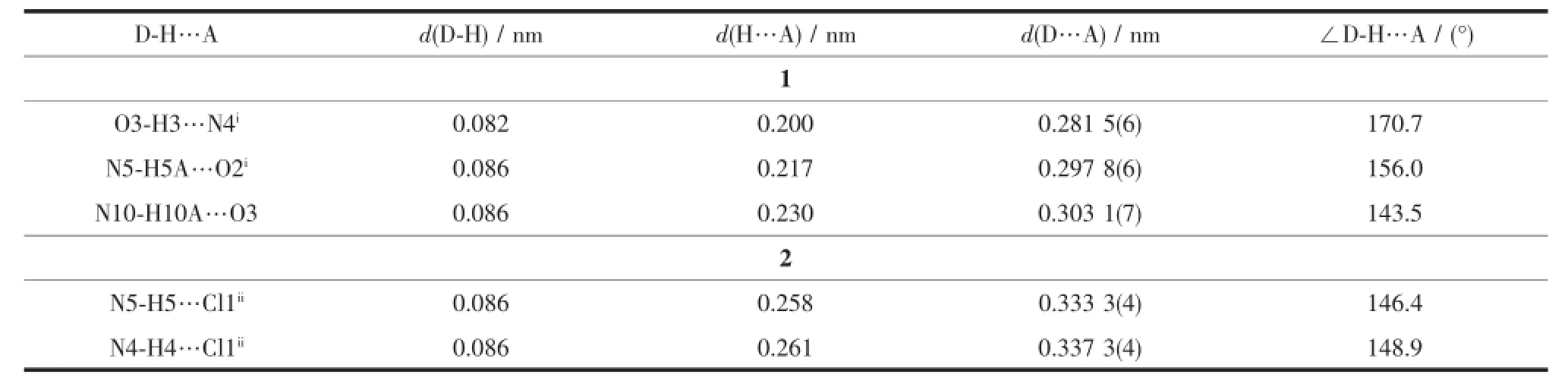
Table 3Hydrogen bonds information for complexes 1 and 2

Fig.1Diamond drawing of 1(a)and 2(b)with 30%thermal ellipsoids;centrosymmetric dimer in 1(c)and 2(d) formed by hydrogen bonding
By contrast,the ratio of the ligand HL and metal is 1∶1,and the semicarbazone acts as a neutral tridentate ligand in complex 2(Fig.1b),which is confirmed by the double bond nature of C8-O1(bond length being 0.123 6(4)nm)[7-8].The Cdion is also coordinated with two chloride ions,thus giving a distorted square pyramidal geometry with τ=0.071 7[7].In the crystal,similar centrosymmetric dimer is formed dueto the intermolecular N-H…Cl hydrogen bonds(Fig. 1d).
2.2 PXRD and IR spectra
ThepowderX-raydiffraction(PXRD)for complexes 1 and 2 are presented in Fig.2,and the experimental pattern is almost same as that of the simulated pattern,except for a little difference about reflectionintensities,whichmanifeststhattwo complexes are the purity phase.

Fig.2PXRD patterns of complexes 1(a)and 2(b)
The IR spectral regions for both complexes are moreorlesssimilarduetothesimilarityin coordination modes of the ligand with the metal center.The ν(C=O)of the free ligand is 1 702 cm-1, while it is disappeared in complex 1,meanwhile,new (N=C-O)stretching vibration absorption is observed at 1 615 cm-1,revealing that the C=O in O=C-N moiety has enolizated and the oxygen atom coordinates to the metal ion[7].By contrast,ν(C=O)of complex 2 shifts to 1 642 cm-1,indicating the coordination of carbonyl group of the neutral ligand HL[8].The ν(C=N-N)bands of the imine group and pyrazine ring in the ligand HL shift to lower frequency values in the complexes, showing that the N atoms of both units take part in the coordination[7-8].It is in accordance with the crystal structure study.
2.3 UV spectra
The UV spectra of HL,complexes 1 and 2 in methanol solution(concentration:1×10-5mol·L-1)were measured at room temperature(Fig.3).The spectra of HL features one main band located around 289 nm (ε=14 355 L·mol-1·cm-1),which could be assigned to characteristic π-π*transition of the pyrazine units[8,11]. The band has almost no change in the spectra of 2(289 nm,ε=14 312 L·mol-1·cm-1),while slightly redshifts to 292 nm(ε=13 041 L·mol-1·cm-1)along with hypochromic effect in the spectra of 1.In addition, complex 1 exhibits another band at 400 nm(ε=4 810 L·mol-1·cm-1),corresponding to the ligand-to-metal charge transfer(LMCT)[8].This indicates that an extended conjugation is formed in anionic ligand after complexation in complex 1.
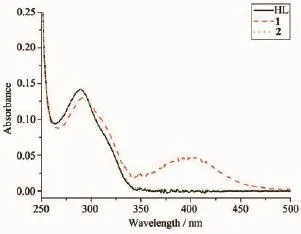
Fig.3UV spectra of the ligand HL,complexes 1 and 2 in CH3OH solution at room temperature
2.4 Thermal analysis
For detecting the thermal stabilities of complexes 1 and 2,thermal gravimetric(TG)analyses were carried out from the room temperature to 800℃with the linear heating rate of 10℃·min-1under argon atmosphere(Fig.4).The first stage occurs with weight lossof7.02%below220℃forcomplex1, contributing to the loss of crystal solvent molecules (Calcd.6.45%).The second and third processes of weight loss appear between 220 to 305℃,305 to 800℃,respectively,considered as the decomposition of the two ligands independently.Complex2 is thermally stable up to about 300℃,indicating there exist no solvent molecules in the complex.The weight lossfrom300to800℃isassignedtothe decomposition of the chloride anions and the organic ligand.The remainders of the complexes 1 and 2 might be the metal oxides because the residue weights (12.52%and26.42%)areagreementwiththe calculated values of 11.64%,and 28.68%,respectively.
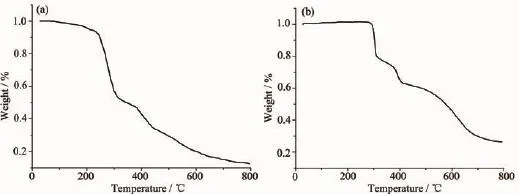
Fig.4TG curves for complexes 1(a)and 2(b)
2.4 EB-DNA binding study by fluorescence spectrum
ItiswellknownthatEBcanintercalate nonspecifically into DNA,which causes it to fluoresce strongly.Competitive binding of other drugs to DNA and EB will result in displacement of bound EB and a decrease in the fluorescence intensity[7,13].As shown in Fig.5,the fluorescence intensities of EB bound to ct-DNA at about 600 nm show remarkable decreasing trends with the increasing concentration of the tested complexes,indicating that some EB molecules are exchanged by the tested complexes.The quenching of EB bound to DNA by the compounds is in agreement with the linear Stern-Volmer equation:I0/I=1+Ksqr[13], where I0and I represent the fluorescence intensities in the absence and presence of quencher,respectively, Ksqis the linear Stern-Volmer quenching constant,r is the ratio of the concentration of quencher and DNA. In the quenching plots of I0/I versus r,Ksqvalues are given by the slopes.The Ksqvalues are 0.527 and 0.415 for complexes 1 and 2,respectively,while that of the ligand HL is tested to be 0.268 in our previouswork[7].The result indicates that interactions of the complexes with DNA are stronger than that of the ligand HL,probably due to the higher rigidityofthe complexes,which is in accordance with the literature[7]. In addition,compared with complex 2,the ratio of metal and ligand is 1∶2 and both ligands are deprotonated and more conjugated in complex 1,thus leading to higher DNA-binding ability.However,the Ksqvalue of complex 1is lower than that of the Cucomplexes with same ligand,namely,[Cu2(L)2Cl2]and [Cu2(L)2(OAc)2](Ksqvalues being 0.723 and 0.780,respectively)[7].This is mainly due to the fact that complex 1has larger sterically hindered effect(octahedron coordination geometry)than the literature complexes (distorted square pyramid coordination geometry)[7], which may prevent the complex 1 molecule from intercalating with DNA.
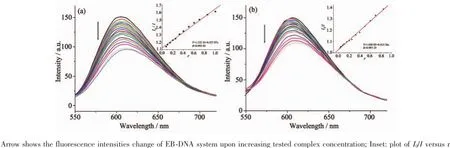
Fig.5Emission spectra of EB-DNA system in the presence of complexes 1(a)and 2(b),respectively
[1]Alagesan L,Bhuvanesh N S P,Dharmaraj N.Dalton Trans., 2013,42:7210-7223
[2]Ye X P,Zhu T F,Wu W N,et al.Inorg.Chem.Commun., 2014,47:60-62
[3]CHEN Yan-Min(陈延民),XIE Qing-Fan(解庆范),LIU Jin-Hua(刘金花),et al.Chinese J.Inorg.Chem.(无机化学学报),2015,31(1):74-80
[4]Singh P,Singh D P,Singh V P.Polyhedron,2014,81:56-65
[5]RecioDespaigneAA,DaCostaFB,PiroOE,etal.Polyhedron, 2012,38:285-290
[6]Chang H Q,Jia L,Xu Z Q,et al.Inorg.Chem.Commun., 2015,57:8-10
[7]LIN Long(林龙),LI Xian-Hong(李先宏),ZHANG Bo(张波), et al.Chinese J.Inorg.Chem.(无机化学学报),2017,33(1): 143-148
[8]MAO Pan-Dong(毛盼东),HAN Xue-Feng(韩学锋),WU Wei-Na(吴伟娜),et al.Chinese J.Inorg.Chem.(无机化学学报),2016,32(1):161-166
[9]Li M X,Zhang L Z,Yang M,et al.Bioorg.Med.Chem. Lett.,2012,22:2418-2433
[10]Li M X,Zhang L Z,Zhang D,et al.Eur.J.Med.Chem., 2011,46:4383-4390
[11]Bourosh P N,Revenko M D,Stratulat E F,et al.Russ.J. Inorg.Chem.,2014,59:545-557
[12]Revenko M D,Bourosh P N,Stratulat E F,et al.Russ.J. Inorg.Chem.,2010,55:1387-1397
[13]MAO Pan-Dong(毛盼东),YAN Ling-Ling(闫玲玲),WANG Wen-Jing(王文静),et al.Chinese J.Inorg.Chem.(无机化学学报),2016,32(3):555-560
[14]Sheldrick G M.SADABS,University of Göttingen,Germany, 1996.
[15]Sheldrick G M.SHELX-97,Program for the Solution and the Refinement of Crystal Structures,University of Göttingen, Germany,1997.
含吡嗪的缩氨基脲配体Ni/Cd配合物的晶体结构及与DNA的相互作用
毛盼东1赵晓雷*,1邵志鹏2李敏2吴伟娜*,1王元1
(1河南理工大学化学化工学院,焦作454000)
(2河南理工大学材料科学与工程学院,焦作454000)
合成配合物[NiL2]·CH3OH·0.5H2O(1)和[Cd(HL)Cl2](2)(HL=3-甲基2-乙酰吡嗪缩4-苯基氨基脲)并通过单晶衍射、元素分析及红外光谱表征其结构。单晶衍射结果表明,配合物1中,Ni离子与2个拥有N2O电子供体的阴离子配体L-配位,配位构型为扭曲的八面体。而配合物2中,Cd离子与1个中性三齿配体HL和2个氯离子配位,拥有扭曲的四方锥配位构型。荧光光谱结果表明,配合物与DNA的相互作用强于配体。
缩氨基脲;镍配合物;镉配合物;吡嗪;晶体结构;DNA相互作用
O614.81+3;O614.24+2
A
1001-4861(2017)05-0890-07
2016-11-26。收修改稿日期:2017-03-28。
10.11862/CJIC.2017.109
国家自然科学基金(No.21001040)、河南省科技厅基础与前沿项目(No.162300410011)、河南省教育厅自然科学基金(No.12B150011,14B150029)和河南省青年骨干教师项目(No.2014GGJS-045)资助。
*通信联系人。E-mail:zhaoxiaolei@hpu.edu.cn;wuwn08@hpu.edu.cn;会员登记号:S06N6704M1112(吴伟娜)。
猜你喜欢
杂志排行
无机化学学报的其它文章
- Three Coordination Complexes Based on an Asymmetric Triazole Derivative Ligand: Syntheses,Structures and Photocatalytic Properties
- Two Tetrapodal Schiff Bases Acting as Colorimetric Sensors for Iron in Environmental Water Samples
- Syntheses,Characterization and Fluorescence Properties Analysis of Two Lanthanide Coordination Polymers Based on Axial Chirality Ligand 2,2′-Dinitro-4,4′-biphenyldicarboxylic Acid
- 磁性UiO-66复合材料的合成及其对水体中硝基酚有机分子的吸附性能
- cis-[(trans-1,2-双(氨甲基)环丁烷) (3-羟基-1,1-环丁烷二羧酸根)合铂(Ⅱ)]的合成和抗癌活性
- 两个具有Sn4O4梯状结构二丁基锡羧酸酯的微波溶剂热合成、结构和体外抗癌活性
