Three-dimensional histology: new visual approaches to morphological changes during neural regeneration
2017-03-30HeiMingLaiHoManNgWutianWu
Hei Ming Lai, Ho Man Ng, Wutian Wu,
1 School of Biomedical Sciences, Li Ka Shing Faculty of Medicine,e University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China
2 State Key Laboratory of Brain and Cognitive Sciences, Li Ka Shing Faculty of Medicine,e University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China
3 Research Center of Reproduction, Development and Growth, Li Ka Shing Faculty of Medicine,e University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China
4 Guangdong‐Hong Kong‐Macau Institute of CNS Regeneration, Jinan University, Guangzhou, Guangdong Province, China
Three-dimensional histology: new visual approaches to morphological changes during neural regeneration
Hei Ming Lai1, Ho Man Ng1, Wutian Wu1,2,3,4,*
1 School of Biomedical Sciences, Li Ka Shing Faculty of Medicine,e University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China
2 State Key Laboratory of Brain and Cognitive Sciences, Li Ka Shing Faculty of Medicine,e University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China
3 Research Center of Reproduction, Development and Growth, Li Ka Shing Faculty of Medicine,e University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China
4 Guangdong‐Hong Kong‐Macau Institute of CNS Regeneration, Jinan University, Guangzhou, Guangdong Province, China
How to cite this article:Lai HM, Ng HM, Wu W (2017)ree-dimensional histology: new visual approaches to morphological changes during neural regeneration. Neural Regen Res 12(1):53-55.
Open access statement:is is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
Three‐dimensional (3D) histology utilizes tissue clearing techniques to turn intact tissues transparent, allowing rapid interrogation of tissue architecture in three dimensions. In this article, we summarized the available tissue clearing methods and classi fi ed them according to their physicochemical principles of op‐eration, which provided a framework for one to choose the best techniques for various research settings. Recent attempts in addressing various questions regarding the degenerating and regenerating nervous system have been promising with the use of 3D histological techniques.
three-dimensional histology; tissue clearing; neuronal morphology; neuronal network
Accepted: 2016-11-10
Introduction
The study of neural tissue architecture and cellular mor‐phology is important in neuroscienti fi c research. Visualizing and understanding the three‐dimensional (3D) spatial re‐lationships of cells and molecules have been di fficult using conventional histology. Serial sections of tissues are often required and this method is time‐ and labor‐intensive, and leads to physical damage and depletion of tissue samples. Advances in optical microscopy and fl uorescent probes have contributed to the emergence of tissue‐clearing methods that allow intact, transparent tissues to be optically sectioned, imaged, and reconstructed in 3Din silico. Here, we briefly introduce the techniques that facilitate the rigorous histolog‐ical evaluation of intact transparent tissues in 3D and better visualization of neuronal tracts and fi bers, which could allow researchers to gain extraordinary insights into the processes of neurodegeneration and neuroregeneration.
As a general rule, aqueous‐basedn‐homogenization (Susaki et al., 2016) best preserves tissue structures and fl uorescence, but the lack of adequate permeabilization makes it incompat‐ible or poorly compatible with immunohistochemistry. More‐over, the transparency of the tissues is not as good as the oth‐er two categories. In delipidation‐facilitated tissue‐clearing methods, the tissues are washed free of lipids in detergents with subsequentn‐homogenization using a suitable aqueous medium (Chung et al., 2013; Susaki et al., 2016).e aqueous media and permeabilization result in good preservation of endogenous fl uorescence and compatibility with immunohis‐
tochemistry, respectively, but they are incompatible with most lipophilic tracers and limited by the long time required for ad‐equate tissue delipidation. In organic solvent‐basedn‐homog‐enization methods, the tissues are dehydrated and immersed in a hydrophobic organic solvent for homogenization (Ertürk et al., 2012).ey are compatible with immunohistochemistry (Renier et al., 2014), but commonly destroy the fluorescent proteins expressed using genetic and viral tools if the pH is not well‐controlled (Schwarz et al., 2015). With the di ff ering char‐acteristics and advantages of di ff erent methods of tissue‐clear‐ing, one size does not fi t all (able 1).us, when choosing a method of tissue‐clearing for speci fi c research questions and goals, a thorough understanding of the physicochemical basis of individual methodologies and their impact on tissues is im‐portant, as it allows the researcher to select the methodology appropriate for the desired application.
able 1 Selected tissue clearing methods

able 1 Selected tissue clearing methods
Summary of selected tissue‐clearing methods classi fi ed according to suggested working mechanisms, advantages, and disadvantages. Note that the list is general and exceptions exist; for example, lipophilic tracers can be made compatible with delipidation‐facilitatedn‐homogenization by using fi xable analogues; SWITCH, a delipidation‐facilitated method, is associated with signi fi cant tissue discoloration and auto fl uorescence; FASTClear is a novel strategy in which tissue‐clearing can be facilitated by delipidation followed by aqueous or organic solvent‐basedn‐homogenization. CLARITY: Clear Lipid‐exchanged, Acrylamide‐hybridized Rigid Imaging/Immunostaining/In situ hybridization‐compatible Tissue hYdrogel; SWITCH: System‐Wide Control of Interaction Time and kinetics of Chemicals; FASTClear: Free‐of‐Acrylamide, SDS‐based Tissue Clearing; BABB: Benzyl Alcohol‐Benzyl Benzoate tissue clearing; 3DISCO: 3‐Dimensional Imaging of Solvent‐Cleared Organs; uDISCO: ultimate 3‐Dimensional Imaging of Solvent‐Cleared Organs.
Aqueous‐basedn‐homogenization Delipdation‐facilitatedn‐homogenization Organic solvent‐basedn‐homogenization Examples ScaleS (Hama et al., 2015), ClearT(Kuwajima et al., 2013) BABB (Schwarz et al., 2015), 3DISCO (Ertürk et al., 2012), uDISCO (Pan et al., 2016), FASTClear (Liu et al., 2016) Unique advantages CLARITY (Chung et al., 2013), CUBIC (Tainaka et al., 2015), SWITCH (Murray et al., 2015), FASTClear (Liu et al., 2016) Compatible with lipophilic tracers and subsequent ultrastructural studies Best results with immunostaining, least tissue discoloration Best for lipid‐rich regions Incompatible with lipophilic tracers, ultrastructural evaluation, signi fi cant tissue discoloration and auto fl uorescence, can be incompatible with fl uorescent proteins Disadvantages Incompatible/poorly compatible with immunostaining, comparatively poor tissue transparency Incompatible with lipophilic tracers, ultrastructural evaluation, can be slow for lipid‐rich regions, comparatively long tissue processing time
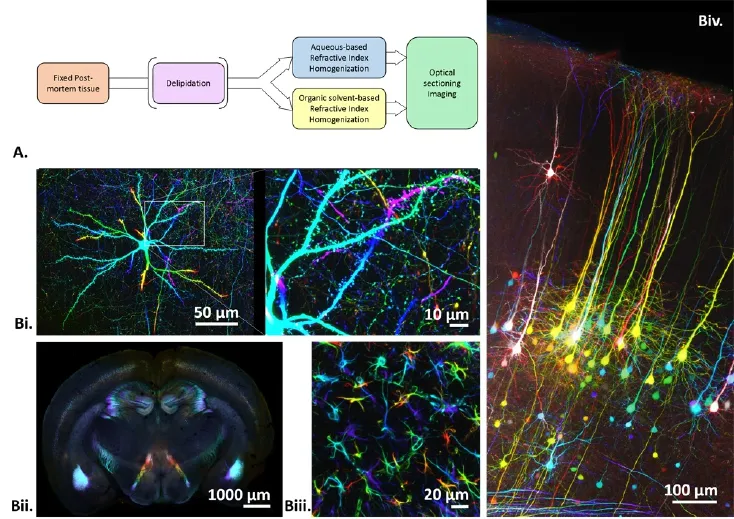
Figure 1 An introduction of 3D histology and some image examples.
Despite the signi fi cant advances in tissue‐clearing, sever‐al methodological challenges remain unresolved. First, the limited diffusion and penetration of antibodies: even with maximal tissue permeabilization by complete delipidation of tissues, the dense location of antigens at shallower depths can rapidly consume antibodies, limiting deep immunos‐taining.is also contributes to di fficulty in the translation of these techniques to human neural tissues, which has been inherently di ffi cult due to the substantially larger tissue vol‐umes and variable sample quality (Lai et al., unpublished ob‐servations). Finally, with the use of ultrafast light‐sheet mi‐croscopic imaging, the volume of data generated commonly lies in the gigabyte to terabyte range, requiring rigorous and robust computational infrastructures for storage and anal‐yses (Susaki et al., 2016). Since these techniques are still in their infancy, they need to be individually optimized by us‐ers in order to obtain the best results for their own projects, emphasizing once again the importance of understanding the working principles underlying these methodologies.
The perfect combination of tissue‐clearing, fluorescent labeling, and optical sectioning microscopy has led to the birth of 3D histology, providing new, exciting, and powerful visualization approaches to the study of the nervous system. Reminiscent of the invention of microtome sectioning, tissue stains, and microscopy that gave birth to histology centuries ago, we envision that the continued methodological innova‐tions in interrogating the brain will achieve new heights in neuroscience research.
Acknowledgments:
Author contributions:All authors contributed to the writing of the manuscript while HML prepared the fi gures and the video.
Con fl icts of interest:None declared.
Supplementary information:Supplementary data associated with this article can be found, in the online version, by visiting www.nrronline.org.
Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Da‐vidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, Pak S, Bernstein H, Ramakrishnan C, Grosenick L, Gradinaru V, Deisseroth K (2013) Structural and molecular interrogation of intact biological systems. Nature 497:332‐337.
Ertürk A, Becker K, Jährling N, Mauch CP, Hojer CD, Egen JG, Hellal F, Bradke F, Sheng M, Dodt HU (2012)ree‐dimensional imaging of solvent‐cleared organs using 3DISCO. Nat Protoc 7:1983‐1995.
Mauch CP, Hellal F, Förstner F, Keck T, Becker K, Jährling N, Ste ff ens H, Richter M, Hübener M, Kramer E, Kirchho ff F, Dodt HU, Bradke F (2011)ree‐dimensional imaging of the unsectioned adult spinal cord to assess axon regeneration and glial responses aer injury. Nat Med 18:166‐171.
Hama H, Hioki H, Namiki K, Hoshida T, Kuroka H, Kurokawa H, Ishidate F, Kaneka T, Akagi T, Saito T, Saido T, Miyawaki A (2015) ScaleS: an optical clearing palette for biological imaging. Nat Neuro‐sci 18:1518‐1529.
Jung Y, Ng JH, Keating CP, Senthil‐Kumar P, Zhao J, Randolph MA, Winograd JM, Evans CL (2014) Comprehensive evaluation of pe‐ripheral nerve regeneration in the acute healing phase using tissue clearing and optical microscopy in a rodent model. PLoS One 9:e94054.
Kuwajima T, Sitko AA, Bhansali P, Jurgens C, Guido W, Mason C (2013) ClearT: a detergent‐ and solvent‐free clearing method for neuronal and non‐neuronal tissue. Development 140:1364‐1368.
Lai HM, Liu AK, Ng WL, DeFelice J, Lee WS, Li H, Li W, Ng HM, Chang RC, Lin B, Wu W, Gentleman SM (2016) Rationalisation and validation of an acrylamide‐free procedure in three‐dimensional his‐tological imaging. PLoS One 11:e0158628.
Liu AK, Lai HM, Chang RC, Gentleman SM (2016) Free‐of‐Acrylamide SDS‐based Tissue Clearing (FASTClear): A novel protocol of tissue clearing for three‐dimensional visualisation of human brain tissues. Neuropathol Appl Neurobiol doi: 10.1111/nan.12361.
Luo X, Yungher B, Park KK (2014) Application of tissue clearing and light sheet fl uorescence microscopy to assess optic nerve regenera‐tion in unsectioned tissue. Methods Mol Biol 1162:209‐217.
Murray E, Cho J, Goodwin D, Ku T, Swaney J, Kim S, Choi H, Park Y, Park J, Hubbert A, McCue M, Vassallo S, Bakh N, Frosch MP, Wedeen VJ, Seung S, Chung K (2015) Simple, scalable proteomic imaging for high‐dimensional pro fi ling of intact systems. Cell 163:1500‐1514.
Pan C, Cai R, Quacquarelli FP, Ghasemigharagoz A, Lourbopoulos A, Matryba P, Plesnila N, Dichgans M, Hellal F, Ertürk A (2016) Shrink‐age‐mediated imaging of entire organs and organisms using uDIS‐CO. Nat Methods 13:859‐867.
Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier‐Lavigne M (2014) iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159:896‐910.
Schwarz MK, Scherbath A, Sprengel R, Engelhardt J,eer P, Giese G (2015) Fluorescent protein‐stabilization and high‐resolution imaging of cleared, intact mouse brains. PLoS One 10:e0124650.
Soderbolm C, Lee D, Dawood A, Carballosa M, Santamaria JA, Benavides FD, Jergova S, Grumbles RM,omas CK, Park KK, Guest JD, Lem‐mon VP, Lee JK, Tsoulfas P (2015) 3D imaging of axons in transparent spinal cords from rodents and nonhuman primates. eNeuro 2.
Susaki EA, Ueda HR (2016) Whole‐body and whole‐organ clearing and imaging techniques with single‐cell resolution: toward organ‐isms‐level systems biology in mammals. Cell Chem Biol 23:137‐157.
Tainaka K, Kubota SI, Suyama TQ, Susaki EA, Perrin D, Ukai‐Tade‐numa M, Ukai H, Ueda HR (2015) Whole‐body imaging with sin‐gle‐cell resolution by tissue decolorization. Cell 159:911‐924
Treweek JB, Gradinaru V (2016) Extracting structural and functional features of widely distributed biological circuits with single cell reso‐lution via tissue clearing and delivery vectors. Curr Opin Biotechnol 40:193‐207.
Wutian Wu, M.D., wtwu@hku.hk.
10.4103/1673-5374.198974
*< class="emphasis_italic">Correspondence to: Wutian Wu, M.D., wtwu@hku.hk.
杂志排行
中国神经再生研究(英文版)的其它文章
- Restoring axonal localization and transport of transmembrane receptors to promote repair within the injured CNS: a critical step in CNS regeneration
- Information for Authors -Neural Regeneration Research
- A new computational approach for modeling diffusion tractography in the brain
- Celebration of the 10thAnniversary of Neural Regeneration Research
- Terapeutic potential of brain-derived neurotrophic factor (BDNF) and a small molecular mimics of BDNF for traumatic brain injury
- Blood microRNAs as potential diagnostic markers for hemorrhagic stroke
