Achievements and prospects of grass pea(Lathyrus sativus L.)improvement for sustainable food production
2016-10-24GirishPrasadDixitAshokKumarPariharAbhishekBohraNarendraPratapSingh
Girish Prasad Dixit,Ashok Kumar Parihar*,,Abhishek Bohra,Narendra Pratap Singh
ICAR-Indian Institute of Pulses Research(IIPR),Kanpur 208024,India
Achievements and prospects of grass pea(Lathyrus sativus L.)improvement for sustainable food production
Girish Prasad Dixit1,Ashok Kumar Parihar*,1,Abhishek Bohra1,Narendra Pratap Singh
ICAR-Indian Institute of Pulses Research(IIPR),Kanpur 208024,India
A R T I C L E I N F O
Article history:
Available online 21 July 2016
Grass pea
Genetic resource
Genetic improvement
Molecular marker
Molecular breeding
Toxin
Grass pea offers an attractive choice for sustainable food production,owing to its intrinsic properties including limited water requirement and drought tolerance.However,low productivity and the presence of a neurotoxin(ODAP)have posed major obstacles to its genetic improvement.Also,biotechnological investments remain limited and the genome is complex and not well understood.Strategies that allow identification of genotypes with reduced ODAP content,coupling of low ODAP content with enhanced yield,and effective seed detoxification methods merit immediate attention.Breeder-friendly genomic tools are being increasingly made available to improve the efficiency of breeding protocols.To this end,the application of next-generation sequencing has provided a means of leveraging the repertoire of genomic resources for this somewhat neglected crop.In this review,we describe progress achieved in Lathyrus genetic improvement.We also explore potential opportunities in Lathyrus research and identify urgent research needs.
©2016 Crop Science Society of China and Institute of Crop Science,CAAS.Production and hosting by Elsevier B.V.This is an open access article under the CC BY-NC-ND license
(http://creativecommons.org/licenses/by-nc-nd/4.0/).
1.Introduction
Grass pea(Lathyrus sativus L.)is a crop of immense economic significance,especially in developing nations including India,Bangladesh,Pakistan,Nepal,and Ethiopia[1-3].It is often broadcast-seeded into standing rice crops one or two weeks before the rice harvest.This allows grass pea to effectively exploit the residual moisture left after the rice harvest[4,5].It is also cultivated in China and in many countries of Europe,the Middle East,and Northern Africa.It serves a variety of purposes including food,feed and fodder,owing in part to its nutritive qualities[4,6-9].Archeological evidence suggests that the domestication of Lathyrus dates to the late Neolithic,and precisely to the Bronze Age[2,9].Prior to domestication,the crop was presumably present as a weed among other pulse crops.However,evidence based on historical records renders the subject of its origin more contentious[10].
In South Asian countries,grass pea is commonly grown for both grain and fodder purposes.However,the crop has gained more importance for use as animal feed than for use as human food.Animal feed from Lathyrus is usually composed of ground or split grain or flour,and is used primarily to feed lactating cattle or other draft animals[11,12].Human diets include Lathyrus as grains that are boiled and then either consumed whole or processed for split dal[13,14].
Grass pea,inherently capable of withstanding temperature extremes,is grown across diverse regions that receive anaverage annual precipitation ranging from 300 to 1500 mm[15,16].In addition to remarkable tolerance to drought[17],Lathyrus has tolerance to excess precipitation and flooding[7,18-20].It has a hardy and penetrating root system suited to a wide range of soil types including very poor soil and heavy clays[20-22].Its notable robustness along with its intrinsic ability to biologically fix atmospheric nitrogen makes grass pea an attractive crop for adverse agricultural conditions[4,18,23].
Nutrient-dense food crops with reduced water demands such as Lathyrus are likely to play a key role in alleviating global malnutrition.However,to date,very limited research efforts have been devoted to improving Lathyrus.The major reason underlying this lack of research effort is the presence of a neurotoxin [β-N-oxalyl-L-α,β-diaminopropionic acid(ODAP)],prolonged consumption of which leads to the neurological disorder lathyrism in humans and domestic animals[24-26].The disease is more pronounced when grass pea forms the dominant component of the diet and accounts for at least 30%of caloric intake for a period of at least three to four months[18,27].Influences of a variety of factors on ODAP accumulation in Lathyrus,including plant growth stage,nutrients,abiotic stresses(drought,salinity,water,and heavy metal)have been comprehensively reviewed by Jiao et al.[28].
2.Implications for human health
As indicated in the previous section,the major health concern associated with grass pea consumption is the neurotoxin ODAP,which is also known as β-N-oxalyl-amino-L-alanine(BOAA)[29,30].Irreparable loss of motor function may result from prolonged consumption of Lathyrus grains[26].Instances of lathyrism have been reported from various parts of India[15,29,31].It was observed that lathyrism could affect anyone consuming a diet consisting of more than 25%grass pea for 3-4 months[15,32].In view of these findings,the sale or storage of Lathyrus has been banned in all states in India except for Chhattisgarh,Maharashtra,and West Bengal under rule 44-A of the Prevention of Food Adulteration Act,1954[33].
In India,Lathyrus cultivation is concentrated mainly in the Chhattisgarh region,where limited cases of human lathyrism areknown.Incontrast,ahigherincidenceofhuman lathyrism has been reported in Rewa division of Madhya Pradesh state,occurring when Lathyrus constituted more than 2/3 of the diet for prolonged periods(3-6 months)[34,35].A survey of the socioeconomic conditions of Lathyrus-growing farmers and their culinary practices was conducted in two villages near Raipur.In these villages,the consumption was related to the size of the farm holdings,with non-farm holding families having greater consumption of Lathyrus[36].
Grass pea holds tremendous potential as a functional food to improve health conditions associated with cardiovascular disease,hypoxia,and hypertension [37-39].Importantly,patents have been granted based on ODAP(as a hemostatic agent)in the USA and China,and an increasing number of therapeuticapplicationsderivedfromLathyrusmaybe developed in coming years[38].Further,as highlighted by Singh and Rao[39],ODAP is increasingly being used for therapeutic purposes owing to its role in the stabilization of hypoxia inducible factor-1(HIF-1).In short,the evolving view of grass pea as a functional food is likely to cause a dramatic shift in the ways pulses and lathyrism are perceived.
3.Economic importance in India
Grass pea is the third most important cool-season pulse crop of India,occupying an area of 0.58 million ha with an annual production of 0.43 million tonnes[14].It is cultivated primarily in Bihar,Madhya Pradesh,Maharashtra,West Bengal,and Chhattisgarh[37].The majority of this acreage(~70%)is shared by Chhatisgarh and the Vidarbha region of Maharashtra,which is a rice-growing region where supplemental irrigation is available only for rice.Consequently,water is not available for subsequent winter crops,making grass pea the only alternative for a crop following rice[40-42].
Grass pea effectively withstands unfavorable conditions including excessive moisture at sowing,which is often followed by moisture stress at advanced growth stages.In fact,grass pea is preferred for cultivation in such areas owing to its hardy nature coupled with its marginal costs of cultivation.In early 1990s,the socioeconomic impact of grass pea consumption was assessed in a random sample of 100 farmers from Raipur,Bilapur and Bastar.This study revealed that almost 60%of the ricegrowersincludedgrasspeaintheircroppingsystem.Mostof the farmers practiced subsistence agriculture with smaller land holdings(of below 5 ha).However,its consumption among non-farmers did not exceed 3%of total food intake.Among pulses,farmers had a preference for chickpea,which accounted for over 35%of total pulse expenses incurred,followed by other pulse crops including pigeonpea(25.3%),blackgram(17.5%),and grass pea(11.2%).The most common use of grass pea was to preparedal,andnearly25%ofconsumersadoptedconventional measures to detoxify grass pea grains before consumption. Considerable awareness was found among rural people about the toxic effects of grass pea consumption.Another study conductedinGondiadistrictinMaharashtrashowedthatnearly 60%ofthepopulationconsumedgrasspeaasapartoftheirdiet;however,the quantity of grass pea consumed per day was reported to be less than 25 g[37].
4.Trends in area and production in India
Given severe legislative control measures imposed by several state governments,acreage under grass pea has declined considerably over the past decades.Although improved varieties containing low amounts of toxin have been developed by the Indian Council of Agricultural Research(ICAR)and associated agricultural institutes,farmers are increasingly shifting towards higher-value crops.Consequently,a continuous reduction has occurred in the area and production of grass pea across India. The national acreage has gradually fallen from 1.67 million ha to 0.58 million ha over the last four decades.A similar trend has been noted in its production,which has declined substantially from 0.84 million to 0.43 million tons over the same time period(http://agricoop.nic.in/).
5.Major production constraints
Grass pea is grown predominantly under rainfed conditions and on marginal and submarginal lands that are generally characterized by poor soil health/fertility.Apart from various agroecological constraints,the crop also encounters a range of biotic stresses including powdery mildew(Erysiphe pisi),rust(Uromyces fabae),downy mildew(Peronospora lathyri-palustris),thrips(Caliothrips indicus)and abiotic factors including moisture and waterlogging stress,which reduce yield potential by 15%-25%[11].Other factors that can adversely affect grass pea production include i)lower productivity of rice fallows,ii)inadequate seed supply of improved and higher-yielding varieties,iii)lack of adoption of novel crop production technologies,iv)inadequate or unbalanced use of fertilizer,v)untimelysowingandlowseedrates,andvi)weedinfestation[43]. Despite results suggesting that the application of micronutrients to deficient soils could be a cost-effective way to increase grass pea production,farmers rarely adopt such practices,owing to their poor economic status[44,45].Inadequate transfer of applicable technologies remains another constraint.
6.Crop management approaches to improve yields
Grass pea is a winter-season crop adapted to areas with arid or semiarid conditions.In the Indian subcontinent,the crop is generally sown in October or November and harvested in February or March[46].Sowing date is largely determined by the time when the monsoon rains end,soil type,and soil moisture profile during October.In some rainfed areas of India such as Madhya Pradesh,Lathyrus is grown as mixed crop with wheat,chickpea,barley and linseed[43,47].In general,the seeds are sown about two weeks after plowing at a density of 40-50 kg ha-1.
Among the various nutrients,phosphorus(P)and molybdenum(Mo)influence the productivity ofpulses.A study aimed at increasing the productivity of Lathyrus under varying levels of P and foliar spraying of Mo was conducted in West Bengal state.Application of P resulted in a marked yield increase. Yield increases were thought to be a consequence of profuse nodulation,withtheresultingincreaseinNfixationinfluencing therateofphotosynthesis.Inaddition,amarkedincreaseinthe seed yield and overall improvement in yield attributes was obtained by foliar spraying with 0.05%Mo[48].As a crop grown under rainfed multiple-cropping systems,Lathyrus influenced soil fertility,especially in terms of organic matter,total N,and available P.Higher soil fertility in the rainfed crop systems was evident from higher yields of green gram after Lathyrus[49].
Inuteracroppingsystems,Lathyrus(cv.Ratan)outperformed chickpea(cv JG-74),lentil(cv JLS-3)and linseed(cv R-552)when grown after medium-duration rice(cv.Mahamaya:130 days). Utera is a relay cropping system that entails broadcasting seeds of a succeeding crop nearly 15 days prior to the harvest of a rice crop,thereby allowing the succeeding crop to efficiently harness residual moisture.Lathyrus yields were higher when seeds were sown under minimal tillage after rice harvest. Application of N:P:K at 80:60:30 kg ha-1to rice and N of 10 kg ha-1to Lathyrus at planting resulted in the greatest yields for both rice and Lathyrus.Leaving 20 cm of rice stubble height resultedinimprovedgrowth and highest yieldsof Lathyrus.Rice varieties Mahamaya(130 days)and Bamleshwari(140 days)are preferred for utera cropping(with Lathyrus)owing to their greater tolerance to biotic and abiotic stresses(drought/ bacterial blight/gall midge)and higher yield potential.Direct drill seeding of Lathyrus at rates of 40 and 60 kg ha-1resulted in similar yields.Seed treatment with Rhizobium provided significantly higher seed yield(1101 kg ha-1)than no Rhizobium(943 kg ha-1).Application of 20 and 30 kg N ha-1resulted in significantlyhigherseedyieldthan0and10 kg N ha-1(Personal communication,R.L.Pandey,2008).

Table 1-Accessions identified for agronomically/economically important traits.
7.Genetic enhancement of Lathryus
7.1.Collection and conservation of genetic resources
Increasing genetic erosion presents a challenge to sustaining Lathyrus genetic diversity.There is a pressing need to collect and conserve the genetic variation present across diverse geographical regions.A systematic collection of grass pea in India was initiated in 1967 in Madhya Pradesh[50].In 1969 germplasm collections were made in different states of India including Bihar,Eastern Uttar Pradesh,West Bengal,and Gujarat,and from tribal areas of Bihar in 1975[40].More than 1000 accessions were collected from Madhya Pradesh during 1989-1991.A set of 24 diverse and determinate land races designated as LSP-1 to LSP-24 was collected during 2000 from Kangra valley,Himachal Pradesh [51].At present,2720 Lathyrus accessions are conserved in collections at the National Bureau of Plant Genetic Resources(NBPGR),NewDelhi(http://www.nbpgr.ernet.in/PGR_Databases.aspx)[52]. Inaddition,activecollectionsarebeingmaintainedat different research stations including IGAU,Raipur,Indian Institute of Pulses Research(IIPR),Kanpur and NBPGR,Akola. Lathyrus accessions displaying important traits are listed in Table 1.
7.2.Estimating genetic variation for ODAP content
Pronounced variation in Lathyrus grain ODAP content was detected by evaluating diverse grass pea accessions.The range of ODAP content varied across studies.Results have included ranges of 0.15%-0.95%among 1128 accessions[1];0.06%-0.71% among1963accessions[52];0.1-2.6%among576accessions[53];0.1%-0.3%among 1500 accessions[54];0.10%-0.78%across 643 accessions[55];0.2%-2.0%across 100 accessions[56],and 0.128%-0.872%across 1187 accessions[43,57].Notably,grass pea germplasm from the Indian subcontinent contained higher ODAP(0.7%-2.4%)than local germplasm(0.02%-1.20%)from the Near East[58].
Studies of the inheritance of ODAP content have produced different conclusions.Analysis of F2populations suggested that ODAP content was inherited both quantitatively and qualitatively[59-62].Furthermore,presence of non-additive effects[60]as well as additive/additive×additive gene effects for ODAP content was observed across other studies[22,63-65]. Other reports have suggested ODAP content to be controlled predominantly by additive genetic variance[50,66].Results based on reciprocal crosses suggested maternal cytoplasmic effectsonODAPconcentration [22].Astudyconducted in southwestern Australia that included genotypes of both L.sativusandLathyrusciceridetectedsignificantgenotypeeffects on ODAP[67].
7.3.Mutation breeding
The slow progress of grass pea genetic improvement may be due largely to an extremely narrow range of genetic variation resulting from self-pollination and interspecific incompatibility[68].For these reasons,improvement strategies focusing on mutation breeding have been explored as a way of creating genetic variation.Mutation breeding in grass pea has led to the generation of several viable diploid mutants with marked alterations in plant characters such as growth habit,branching,stem and internodes,leaflets,stipules,flower and pod characters,and seed traits[17].Examples include dwarf mutants dwf1,dwf2,and dwf3[69,70].Similarly,branching mutant 1(brm1),brm2 and profusely branched mutant(PBM)were recovered from varieties“BioR-231”and“Hoogly Local”.A non-winged internode mutant(NWIM)with erect,determinate,and semidwarf habit was developed from the variety BioR-231. Interestingly,the mutant also showed an increased number of primary and secondary branches,along with higher grain yield and low seed ODAP content[71].Similarly,a fasciated mutant(250 Gy)was detected,characterized by a broad,strap-like flattened stem,clustered canopy leaves,and reduced yield[72].Mutations affecting leaflet pattern have also been detected by various researchers[68,70,73-75].Interestingly,mutants(CELM)with erect leaflets displayed radiation use efficiency(RUE)and higher biomass accumulation[73].
Mutant lines derived from varieties“BioR-231”,“BioL-203”,and“Nirmal”showed extensive variation in the flower(corolla)[75].Mutants with pale-violet(pvfm)and white flower(wfm)showed high grain-yielding potential[17].As with flower color,a broader range of seed coat variations was developedbymutagenesis[70,76,77].Mutantsincluding black-mosaic seed coat(BSCM)and white non-mosaic seed coat(WSCM)have significantly higher grain yields and lower seed ODAP content than wild type plants[17].The exposed stigma noted in the malformed flower mutation(mfl)could substantially increase pollinator activity[68,78].Similarly,mutants with increased number of pedicels and improved podding capacity were isolated from BioL-203,Nirmal,LSD-3,P-24/3,BioR-231,Hoogly Local,and two local varieties from the eastern Himalayas[17].
Similarly to morphological changes,chromosomal alterations including translocations have been induced in grass pea by mutagenesis[79,80].Recently,a mutant(rlfL-1)was isolated from M2progeny of EMS-treated BioL-212,exhibiting marked deviations in karyology[81].Mutants have been developed that show enhanced salt tolerance[82,83],resulting from an increase in the activities of reactive oxygen species(ROS)-scavenging enzymes including superoxide dismutase and ascorbate peroxidase.
Mutagenic treatments have introduced marked biochemical changes in grass pea,a majority of which have been reported to contribute directly or indirectly to plant defense systems. For example,a glutathione(GSH)-deficient mutant(gshl-1)was isolated from gamma ray-treated M2progeny of the genotypeBioL-212.Examinationofthismutantunder normal and cadmium(Cd)-stressed conditions revealed its greater sensitivity to Cd[84].By contrast,enhanced tolerance to salt and metal toxicity was obtained through induced mutagenesis[83,85].One such mutant,dwf1,showed a 2.5-fold increase in foliar GSH content and normal growth under Cd stress[69,86].
An EMS-induced mutant,rlfL-1,was characterized as showing elevated rates of cell division and cell growth.Similarly,mutants including gshl-1,an ascorbate(AsA)-deficient mutant(asfl-1),and a GSH-overproducing mutant were investigated to assess the role of arsenic(As)on wilt tolerance[85].In addition to underscoring the effect of As on PAL activity,the study suggested a role for H2O2level and GSH redox during pathogenesis[85].
Gamma-raymutagenesisproducedanasfL-1mutant containing only 42%leaf and 20%root ascorbate content relative to the parent genotype BioR-231.The results pointed to the possible occurrence of a rearrangement event involving antioxidantdefensemachineryinasfL-1thateffectively mitigated the adverse effect of ascorbate deficiency and permitted survival under salt-stress conditions[87].Two flavonoid-deficient mutants,fldL-1 and fldL-2,were established asaproductofEMSmutagenesisandleafflavonoidcontentwas found to be reduced by up to 20%in both mutants relative to wild-type genotypes[88].
7.4.Screening against diseases and pests
Thrips are a pest of high economic concern for grass pea cultivation in India.Screening of 56 accessions against thrips atRaipur revealed two genotypes,RLK-273-1 and RLK-273-3,with moderate resistance[57].Similarly,genotypes RLK-1,RLK-281,RLK-617,and RPL-26 were reported to be thrips-tolerant by Asthana and Dixit[40].In addition to pest problems,powdery mildew and downy mildew represent two major biotic factors severely effecting grass pea[11].A three-year study performed under condition of natural infection detected promising lines with no visible symptoms of downy mildew[89].Lines showing moderate resistance to powdery mildew included RPLK-26 and RL-21[90].On the basis of field-based screening,IPLy2-10,RPLK-26,JRL-41,andRLS-2werefoundtobetoleranttopowdery mildew[25].Screening 96 lines with Cercospora pisi sativae f.sp. Lathyri misha revealed eight lines with exceptional resistance or immunity[91].Germplasm with resistance to downy mildew has also been reported[40,57].Vaz Patto and Rubiales[92]screened a set of 50 Lathyrus accessions for rust disease and established the presence of high resistance in all accessions against Uromyces viciae-fabae and Uromyces ciceris arietini,and a partialresistancetoUromycespisi.Detailedhistologicalevidence wasprovidedtoexplaintheresistanceobservedagainsttherust disease.Similarly,a survey of 151 L.ciceri accessions for their response to broomrape(Orobanche crenata)led the authors to advocate to harnessing the joint benefits of both genetic resistance and escape mechanisms to develop genotypes with stable resistance against broomrape[93].
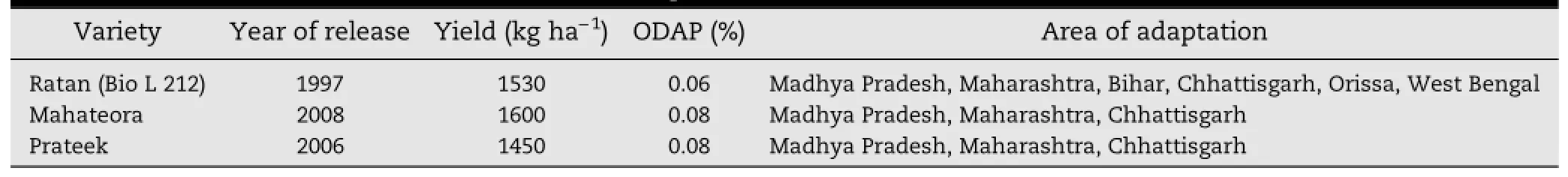
Table 2-Low-ODAP varieties and their areas of adaptation.
7.5.Varietal improvement The first phase of Lathyrus crop improvement in India lasted for almost 20 years(1940-1960)and focused on isolating single plant progenies with substantial yield superiority[25]. As a result,several cultivars were developed and recommended for cultivation.This set of improved cultivars included BR13,LC76(Bihar);T12(Gujarat)No.91/No.11(Madhya Pradesh)and B1/B19(West Bengal).
The second phase(1974-1990)involved development of improved varieties that were relatively free of ODAP.In this phase research was focused on estimating the varying levels of ODAP,and effective breeding protocols were adopted to eliminate or reduce the content of this toxin.
The extensive testing and evaluation led to the release of a low-ODAP(0.2%)variety,Pusa 24,that was particularly adapted to upland cultivation[90,94].Subsequent research efforts led to the development of six additional varieties(LSD1,LSD2,LSD3,LSD6,Pusa-305,and Selection 1276)with low ODAP content(up to 0.2%).Of these,two varieties(LSD1 and LSD2)were recommended for upland cultivation,while the remaining four(LSD3,LSD6,Pusa 305,and Sel.1276)were found to be suitable for rice fallows[25,95].
In the third phase(1990s),several varieties and lines were developed that combined low ODAP with high yield potential and resistance to a variety of biotic and abiotic stresses.For example,BioR-202,BioL-203,BioL-212,BioR-231,and BioL-208 had enhanced yield and high harvest index.As shown in Table 2,BioL-212 was identified and released as“Ratan”in 1997 for cultivation in the North East Plain Zone(NEPZ)and Central Zone(CZ)in India[25,57,96].Later,two varieties were developed,Prateek(LS 157-14)and Mahateora(RLS 4595),that had very low ODAP content(<0.1%)and yield up to 1.5 t ha-1[2].
7.6.Prebreeding and distant hybridization
Genetic improvement of crops having a narrow genetic base may require the use of desirable alleles from outside the primary gene pool. ICARDA has collected and conserved nearly 1555 accessions representing 45 wild Lathyrus species from more than 40 countries [2,97]. Screening of wild grass pea for ODAP content has shown that the zero level is virtually absent in the gene pool [98-100]. The lowest ODAP content has been recorded in L. cicera, followed by L. sativus and Lathyrus ochrus [98-100]. Similarly, ODAP content was found to vary between 0.07% and 0.51% across 142 accessions of L. cicera [2].
A toxin-free gene identified in Lathyrus tingitanus can be deployed in breeding to develop varieties with low levels of toxin[101].Wild species including L.ochrus,Lathyrus clymenum[102],and L.cicera[100,103-105]are resistant to broomrape,a trait that is not present in the cultivated gene pool.In addition to its reduced ODAP content,L.cicera may be a promising source of other important agronomic traits including earliness and cold tolerance[105].
There have been several successful instances of the development of interspecific and wide crosses with Lathyrus. Viable seeds were recovered from wide crosses involving L.sativus and two wild species,L.cicera and Lathyrus amphicarpus[106,107].Yunus[108]attemptedinterspecifichybridizationusing 11 wild species and L.sativus,but only a L.cicera×L.amphicarpus cross yielded viable seeds.Successful interspecific hybrids were alsoobtainedfrom the crosses Lathyrusannuus×Lathyrus hierosolymilanusandLathyrusodoratus×Lathyrusbelinenesis[109-111];Lathyrus hirsutus×L.odoratus[107,109,112];Lathyrus articulatus×LathyrusclymenusandL.articulatus×L.ochrus[112,113];L.cicera×Lathyrus blepharicarpus,L.cicera×Lathyrus gorgoni,L.cicera×Lathyrus marmoratus,L.cicera×L.pseudocicera,L.gorgoni×L.pseudocicera,and L.marmoratus×L.blepharicarpus[97,109,114];and Lathyrus rotundifolius×Lathyrus tuberosus and Lathyrus sylvestris×Lathyrus latifolius[97,106,115].The results of interspecific hybridization in grass pea suggest that the identification and transfer of desirable traits from exotic and wild germplasm offer many opportunities for Lathyrus improvement,especially for readily crossable species including L.cicera and L.amphicarpus.Biotechnological tools including tissue culture techniques may also be employed to overcome strong reproductive barriers among different species[116].
8.Genomic research in Lathyrus
Recent reports have documented genomics tools in grass pea for initiating genomics-based crop improvement[117].Genetic diversity in Lathyrus has been detected using diverse molecular markers including restriction fragment length polymorphism(RFLP),random amplified polymorphic DNA(RAPD),and amplified fragment length polymorphism(AFLP)[118-121].
An analysis of 53 Lathyrus species,three Vicia species,and a single variety of Pisum sativum using internal transcribed spacer(ITS),nuclear ribosomal and chloroplast(cp)sequence-specific DNA markers supported an existing taxonomic classification based on morphological traits[122].A limited number(178)of expressed sequenced tags(ESTs)are available at the National Center for Biotechnology Information(NCBI)GenBank for L.sativus.However,many more ESTs(8702)are available from L.odoratus(http://www.ncbi.nlm.nih.gov/genbank/dbest/ dbest_summary/).An initial set of simple sequence repeat(SSR)markers comprising 20 SSRs was developed using an in silico survey[123].The transferability of seven Medicago truncatula EST-SSRs was evaluated across 19 accessions belonging to 11 different genera[124].Seven SSRs were validated across four diverse accessions of Lathyrus(L.sativus,L.cicera,L.ochrus,L. tingitanus)and P.sativum[125].More recently,genotyping of 176 accessions with EST-SSRs revealed a total of 51 alleles with an average gene diversity of 0.43,and the two subpopulations were recovered using a model-based population structure analysis with authors predicting gene flow among the accessions across the geographical regions in India[126].
Recently,19 primer-pairs were designed by surveying an EST databaseofL.sativus[127].ThreehundredEST-SSRmarkerswere screened across 24 grass pea accessions to identify 44 polymorphic loci[128].In addition to L.sativus-specific DNA markers,24 EST-SSRs were chosen to analyze transferability of Medicago truncatula-specificmarkerinthreelegumespecies [129]. Ponnaiah et al.[130]also reported the development of seven Lathyrus-specific EST-SSR markers.In addition to SSRs,156 intron-targeted amplified polymorphic(ITAP)markers from M.truncatula and field pea and DNA markers specific to defense-associated genes in field pea were also shown to have utility for Lathyrus.Cleaved amplified polymorphic sequence(CAPS)and derived-CAPS(dCAPS)were also developed for Lathyrus[131].Based on a linkage analysis of 75 markers including RAPD,morphological and isozyme markers,map locations could be determined for 69 markers on 14 linkage groups(LGs)covering 898 cM of the genome[132].Similarly,mappingof64markersincludingRAPD and sequencetagged site(STS)/CAPS led to the establishment of nine LGs that collectively spanned 803 cM of the genome[133](reviewed in[117]).
Recently,next-generation sequencing(NGS)platforms were usedtodevelop50,000SSRmarkersforgrasspea[134].Later,30of these high-throughput SSRs were employed to analyze 266 Lathyrus accessions and 17 relatives from Africa,Europe,Asia and ICARDA[135].The possibility of gene flow between the European and African accessions,indicated by the population structure analysis,was further supported by unweighted pair group method with arithmetic mean(UPGMA)-based cluster analysis and principal component analysis(PCA).Recently,RNAseq analysis of rust-responsive grass pea genotypes enabled construction of a reference transcriptome assembly with 134,914 contigs[136].Similarly,coupling Illumina technology with serial analysis of gene expression(SAGE)analysis yielded a set of 738UniTagsexpresseddifferentiallyintheleavesofL.sativusasa responsetoAscochytalathyriinoculation[137].Functional categorization associated these genes with processes involved in “biotic/abiotic stress”,“cell metabolism”,and“hormone signaling”.Given the observation that only peroxidase showed differential expression,the authors proposed that a ROS burst might make only a minor contribution to resistance against A. lathyri.These modern molecular tools represent powerful public resources for genome mapping and molecular breeding in grass pea.
9.Future opportunities and research needs
Substantial regions of rice fallows are scattered across India. The location,extent(11.65 million ha),and gross environmental conditions of these areas have been determined using publicly available databases and a geographic information system(GIS)approach.These rice fallows permit cultivation of an additional crop that can efficiently use moisture retained in the soil.Smaller landholdings constitute an enormous,albeit underused,resource for poor farmers who remain confined to subsistence farming,and have restricted opportunities for enterprise and income diversification.Growing a crop,especially a legume crop such as Lathyrus,after rice would have beneficial effects on soil fertility and soil health.
Similarly,in the Eastern Indo-Gangetic plains a sizeable area is monocropped using medium-and long-duration varieties of rice.The lack of irrigation water and delay in vacating the rice field does not normally permit double cropping.The top soil layer generally dries out at the time of rice harvesting and planting a post-rainy season crop is not feasible.Under such conditions,relay cropping of Lathyrus,especially in medium-deep soils,could transform these monocropped areas into double-cropping areas,increasing the productivity and sustainability of the agricultural system. However,such expansions require the development and promotion of low-toxin varieties that are suited to relay cropping and mechanized farming.
Below are listed areas of investigation that deserve immediate attention:
·Large-scale collection,conservation,and evaluation of germplasm from unexplored regions.
·Accelerated development of low-or near-zero-level ODAP genotypes.
·Breeding for increased forage and fodder production.
·Improved understanding of drought-tolerance mechanisms.
·Identification of sources of resistance against biotic stresses.
·Identificationandgeneticmanipulationofenzymesresponsible for ODAP production.
·Increasing accuracy and throughput of ODAP detoxification method.
·Refinement in production technologies used for utera cultivation.
·Generationofsaturatedlinkagemapsandothercost-effective genomic tools.
·Implementing newly developed genomic resources in existing breeding protocols to accelerate efficient development of new varieties.
We consider that meeting the above-mentioned research priorities will help not only to improve grass pea yield but also to introduce this crop to areas where it is not currently produced.
R E F E R E N C E S
[1]S.Kumar,G.Bejiga,S.Ahmed,H.Nakkoul,A.Sarker,Genetic improvement of grass pea for low neurotoxin(ODAP)content,Food Chem.Toxicol.49(2011)589-600.
[2]S.Kumar,P.Gupta,S.Barpete,A.Sarker,A.Amri,P.N. Mathur,M.Baum,Grass Pea,in:M.Singh,H.D.Upadhyaya,I.S.Bisht(Eds.),Genetic and Genomic Resources of Grain Legume Improvement,Elsevier,Oxford,United Kingdom 2013,pp.269-292.
[3]R.Tripp,W.Van der Heide,The erosion of crop genetic diversity:challenges,strategies and uncertainties,Nat. Resour.Perspect.7(1996).
[4]M.Joshi,Status of Grass Pea(Lathyrus sativus L.)Genetic Resources in Nepal,in:P.N.Mathur,V.R.Rao,R.K.Arora(Eds.),Lathyrus Genetic Resources Network:Proceedings of a IPGRI-ICARDA-ICAR Regional Working Group Meeting,December 8-10,1997,National Bureau of Plant Genetic Resources,New Delhi 1998,pp.22-29(IPGRI Office for South Asia,New Delhi,India).
[5]M.P.Bharati,Status of Lathyrus sativus among Grain Legumes in Nepal,in:A.K.Kaul,D.Comber(Eds.),Lathyrus and Lathyrism,Third World Medical Research Foundation,New York,USA 1986,pp.142-145.
[6]A.K.Biswas,Induced mutation in grass pea(Lathyrus sativus L.),in:S.J.Ochatt,S.M.Jain(Eds.),Underutilized and Neglected Crops,Herbs and Spices,Science Press,Enfield,USA 2007,pp.29-39.
[7]M.A.Malek,Genetic Resources of Grass Pea(Lathyrus sativus L.)in Bangladesh,in:P.N.Mathur,V.R.Rao,R.K.Arora(Eds.),Lathyrus Genetic Resources Network:Proceedings of a IPGRI-ICARDA-ICAR Regional Working Group Meeting,December 8-10,1997,National Bureau of Plant Genetic Resources,New Delhi 1998,pp.1-6(IPGRI Office for South Asia,New Delhi,India).
[8]J.Smartt,Grain Legumes:Evolution and Genetic Resources,Cambridge University Press,Cambridge,1990.
[9]Y.Mahler-Slasky,M.E.Kislev,Lathyrus consumption in late Bronze and Iron Age sites in Israel:an Aegean affinity,J. Archaeol.Sci.37(2010)2477-2485.
[10]W.Erskine,J.Smartt,F.J.Muehlbauer,Mimicry of lentil and the domestication of common vetch and grass pea,Econ. Bot.48(1994)326-332.
[11]C.G.Campbell,Grass Pea(Lathyrus sativus L.),Promoting the Conservation and Use of Underutilized and Neglected Crops,Institute of Plant Genetic and Crop plant Research. Gatersleben/International Plant Genetic Resources Institute,Rome,Italy,1997.
[12]I.Khawaja,H.I.T.Khawaja,I.Ullah,N.U.Raja,A.M.Khushk,LathyrisminPakistan:apreliminarysurvey,in:H.K.M.Yusuf,F. Lambein(Eds.),LathyrussativusandHumanLathyrism:Progress and Prospects:Proceedings of the Second International Colloquium on Lathyrus/Lathyrism,December 10-12,1993,Dhaka,University of Dhaka,Dhaka 1995,pp.55-62.
[13]S.S.Deshpande,C.G.Campbell,Genotype variations in BOAA,condensed tannins,phenolics and enzyme inhibition in grass pea(Lathyrus sativus),Can.J.Plant Sci.72(1992)1037-1047.
[14]A.K.Kaul,M.Q.Islam,A.Hamid,Screening of Lathyrus germplasm of Bangladesh for BOAA content and some agronomic characters,in:A.K.Kaul,D.Combes(Eds.),Lathyrus and Lathyrism,Third World Medical Research Foundation,New York 1986,pp.130-141.
[15]A.N.Asthana,Grass pea cultivation in problem areas: present approaches,in:R.K.Arora,P.N.Mathur,K.W.Riley,Y.Adham(Eds.),Lathyrus Genetic Resources in Asia: Proceedings of a Regional Workshop,December 27-29,1995,Indira Gandhi Agricultural University,Raipur,India 1996,pp.43-48(IPGRI Office for South Asia,New Delhi).
[16]R.K.Arora,P.N.Mathur,K.W.Riley,Y.Adham,Lathyrus Genetic Resources in Asia:Proceedings of a Regional Workshop,December 27-29,1995,Indira Gandhi Agricultural University,Raipur,India,1996(IPGRI Office for South Asia,New Delhi).
[17]D.Talukdar,Recent progress on genetic analysis of novel mutants and aneuploid research in grass pea(Lathyrus sativus L.),Afr,J.Agric.Res.4(2009)1549-1559.
[18]C.G.Campbell,R.B.Mehra,S.K.Agrawal,Y.Z.Chen,A.M.A.El Ali,H.I.T.Khawaja,C.R.Yadav,J.Toy,W.A.Araya,Current status and future strategy in breeding grass pea(Lathyrus sativus),Euphytica 73(1994)167-175.
[19]S.K.Sinha,Food Legumes:Distribution,Adaptability and Biology of Yield,Food and Agriculture Organization of United Nations,Rome,1977.
[20]D.Girma,L.Korbu,Genetic improvement of grass pea(Lathyrus sativus)in Ethiopia:an unfulfilled promise,Plant Breed.131(2012)231-236.
[21]A.K.Parihar,G.P.Dixit,D.Singh,Multivariate analysis of various agronomic traits in grass pea(Lathyrus spp.)germplasm,Indian J.Agric.Sci.83(2013)570-575.
[22]A.M.Abd-El-Moneim,B.Van Dorrestein,M.Baum,J.Ryan,G. Bejiga,Role of ICARDA in improving the nutritional quality and yield potential of grasspea(Lathyrus sativus L.),for subsistence farmers in dry areas,Lathyrus Lathyrism Newsl. 2(2001)55-58.
[23]I.P.S.Ahlawat,A.Singh,C.S.Saraf,Effects of winter legumes on the nitrogen economy and productivity of succeeding cereals,Exp.Agric.17(1981)57-62.
[24]M.T.Jackson,A.G.Yunus,Variation in the grass pea(Lathyrus sativus L.)and wild species,Euphytica 33(1984)549-559.
[25]P.L.Gautam,I.P.Singh,J.L.Karihaloo,Need for a crop network on Lathyrus genetic resources for conservation and use,in:P.N. Mathur,V.R.Rao,R.K.Arora(Eds.),Lathyrus Genetic Resources Network:Proceedings of a IPGRI-ICARDA-ICAR Regional Working Group Meeting,December 8-10,1997,National Bureau of Plant Genetic Resources,New Delhi 1998,pp.15-21(IPGRI Office for South Asia,New Delhi,India).
[26]M.C.Vaz Patto,D.Rubiales,Lathyrus diversity:available resources with relevance to crop improvement-L.sativus and L.cicera as case studies,Ann.Bot.113(2014)895-908.
[27]J.Kumar,Utilization of Lathyrus,in:P.N.Mathur,V.R.Rao,R.K.Arora(Eds.),Lathyrus Genetic Resources Network: Proceedings of a IPGRI-ICARDA-ICAR Regional Working Group Meeting,December 8-10,1997,National Bureau of Plant Genetic Resources,New Delhi 1998,pp.57-59(IPGRI Office for South Asia,New Delhi,India).
[28]C.J.Jiao,J.L.Jiang,L.M.Ke,W.Cheng,F.M.Li,Z.X.Li,C.Y. Wang,Factors affecting beta-ODAP content in Lathyrus sativus and their possible physiological mechanisms,Food Chem.Toxicol.49(2011)543-549.
[29]V.V.S.Murti,T.R.Seshadri,T.A.Venkitasubramanian,Neurotoxic compound of the seeds of Lathyrus sativus,Phytochemistry 3(1964)73-78.
[30]S.L.N.Rao,P.R.Adiga,P.S.Sharma,Isolation and characterization of b-Noxalyl-L-a,b-diaminopropionic acid,a neurotoxin from seeds of Lathyrus sativus,Biochemistry 3(1964)432-436.
[31]K.S.Mani,S.Sriramachari,S.L.N.Rao,P.S.Sharma,Experimental neurolathyrism in monkeys,Indian J.Med. Res.59(1971)880.
[32]S.L.N.Rao,K.Malathi,P.S.Sharma,Lathyrism,World Rev. Nutr.Diet.10(1969)214-238.
[33]S.Dixit,S.K.Khanna,M.Das,Non-uniform implementation of ban on Lathyrus cultivation in Indian states leading to unwarranted exposure to consumers,Curr.Sci.94(2008)570-571.
[34]K.T.Ganapathy,M.P.Dwivedi,Studies on Clinical Epidemiology of Lathyrism,Lathyrism Enquiry Field Unit,Indian Council of Medical Research,Gandhi Memorial Hospital,Rewa,Madhya Pradesh,1961.
[35]D.N.Roy,The neurotoxic disease lathyrism,Natl Med.J. India 1(1988)70-80.
[36]R.L.Pandey,O.P.Kashyap,Studies on socio-economic strata and Lathyrus consumption in Rural Madhya Pradesh,in: H.K.M.Yusuf,F.Lambein(Eds.),Lathyrus sativus and Human Lathyrism:Progress and Prospects:Proceedings of the Second International Colloquium on Lathyrus/Lathyrism,December 10-12,1993,Dhaka.University of Dhaka,Dhaka 1995,pp.47-50.
[37]A.L.Khandare,J.J.Babu,M.Ankulu,N.Aparna,A.Shirfule,G.S.Rao,Grass pea consumption&present scenario of neurolathyrism in Maharashtra state of India,Indian J.Med. Res.140(2014)96-101.
[38]S.L.Rao,A look at the brighter facets of β-N-oxalyl-L-α,β-diaminopropionic acid,homoarginine and the grass pea,Food Chem.Toxicol.49(2011)620-622.
[39]S.S.Singh,S.L.N.Rao,Lessons from neurolathyrism:a disease of the past&the future of Lathyrus sativus(Khesari dal),Indian J.Med.Res.138(2013)32-37.
[40]A.N.Asthana,G.P.Dixit,Utilization of genetic resources in Lathyrus,in:P.N.Mathur,V.R.Rao,R.K.Arora(Eds.),Lathyrus Genetic Resources Network:Proceedings of a IPGRI-ICARDA-ICAR Regional Working Group Meeting,December 8-10,1997,National Bureau of Plant Genetic Resources,New Delhi 1998,pp.64-70(IPGRI Office for South Asia,New Delhi,India).
[41]I.P.Singh,K.P.S.Chandel,Lathyrus germplasm resources at NBPGR,India Introduction and Evaluation,in:R.K.Arora,P.N.Mathur,K.W.Riley,Y.Adham(Eds.),Lathyrus Genetic Resources in Asia:Proceedings of a Regional Workshop,December 27-29,1995,Indira Gandhi Agricultural University,Raipur,India 1996,pp.53-57(IPGRI Office for South Asia,New Delhi,India).
[42]V.K.Yadav,S.L.Mehta,Lathyrus sativus:a future pulse crop free of neurotoxin,Curr.Sci.68(1995)288-292.
[43]R.L.Pandey,M.W.Chitale,R.N.Sharma,N.Rastogi,Status of Lathyrus research in India,in:R.K.Arora,P.N.Mathur,K.W. Riley,Y.Adham(Eds.),Lathyrus Genetic Resources in Asia: Proceedings of a Regional Workshop,December 27-29,1995,Indira Gandhi Agricultural University,Raipur,India 1996,pp.45-52(IPGRI Office for South Asia,New Delhi,India).
[44]S.S.Baghel,A.R.S.Sastri,A.K.Geda,Lathyrus cultivation in Chhattisgarh region of Central India,in:R.K.Arora,P.N. Mathur,K.W.Riley,Y.Adham(Eds.),Lathyrus Genetic Resources in Asia,International Plant Genetic Resources Institute,Rome 1995,pp.139-142.
[45]S.L.Mehta,Plant biotechnology for removal of ODAP from Lathyrus,in:R.T.Hainranot,F.Lambein(Eds.),Lathyrus and Lathyrism,A Decade of Progress,University of Ghent,Belgium 1997,pp.103-104.
[46]C.D.M.Sarwar,A.Sarkar,A.N.M.M.Murshed,M.A.Malik,Variation in natural population of grass pea,in:H.K.M. Yusuf,F.Lambein(Eds.),Lathyrus sativus and Human Lathyrism:Progress and Prospects,Proceedings Second International Colloq.Lathyrus/Lathyrism,Dhaka,10-12 December 1993,University of Dhaka,Dhaka 1995,pp.161-164.
[47]R.L.Pandey,M.W.Chitale,R.N.Sharma,O.P.Kashyap,S.K. Agrawal,A.K.Geda,H.K.Chandrakar,K.K.Agrawal,Catalogue on Grass Pea(Lathyrus sativus L.)Germplasm,Department of Plant Breeding&Genetics,Indira Gandhi Agricultural University,Raipur,India,1995,p 60.
[48]R.K.Sarkar,B.Biswas,G.C.Malik,Productivity of grass pea(Lathyrus sativus L.)under different levels of phosphorus and foliar spray of molybdenum,Lathyrus Lathyrism Newsl.3(2003)36-37.
[49]N.R.Das,Lathyrus sativus In rainfed multiple cropping systems in West Bengal,India-a review,Lathyrus Lathyrism Newsl.1(2000)25-27.
[50]R.B.Mehra,D.B.Raju,K.Himabindu,Evaluation and utilization of Lathyrus sativus collection in India,in:R.K. Arora,P.N.Mathur,K.W.Riley,Y.Adham(Eds.),Lathyrus Genetic Resources in Asia:Proceedings of a Regional Workshop,December 27-29,1995,Indira Gandhi Agricultural University,Raipur,India 1996,pp.37-43(IPGRI Office for South Asia,New Delhi,India).
[51]V.Kumari,Stablegenotypesofgrasspeaformidhillconditions of Himachal Pradesh,Indian J.Genet.60(2000)399-402.
[52]R.L.Pandey,O.P.Kashyap,R.N.Sharma,H.C.Nanda,A.K. Geda,S.Nair,Catalogue on Grass Pea(Lathyrus sativus L.)Germplasm,Indira Gandhi Krishi Vishwavidyalaya,Raipur,India,2008.
[53]V.Nagarajan,C.Gopalan,Variation in the neurotoxin(b-N oxalyl amino alanine)content in Lathyrus sativus samples from Madhya Pradesh,Indian J.Med.Res.56(1968)95-99.
[54]L.M.Jeswani,B.M.Lal,S.Prakash,Studies on the development of low neurotoxin(B-N-oxayl amino alanine lines in Khesari(L.sativus L.),Curr.Sci.39(1970)518-519.
[55]P.I.N.Somayajulu,G.K.Barat,S.Prakash,B.K.Mishra,V.C. Shrivastava,Breeding of low toxin containing varieties of Lathyrus sativus,Proc.Nutr.Soc.India 19(1975)35-39.
[56]C.Leakey,Khesari dal-the poisonous pea,Appropr. Technol.6(1979)15-16.
[57]R.L.Pandey,R.N.Sharma,M.W.Chitale,Status of Lathyrus genetic resources in India,in:P.N.Mathur,V.R.Rao,R.K. Arora(Eds.),Lathyrus Genetic Resources Network: Proceedings of a IPGRI-ICARDA-ICAR Regional Working Group Meeting,December 8-10,1997,National Bureau of Plant Genetic Resources,New Delhi 1998,pp.7-14(IPGRI Office for South Asia,New Delhi,India).
[58]A.M.Abd-El-Moneim,B.Van-Dorrestein,M.Baum,W. Mulugeta,Improving the nutritional quality and yield potential of grass pea(Lathyrus sativus L.),Food Nutr.Bull.21(2000)493-496.
[59]B.S.Dahiya,Genetics and stability analysis in grass pea(L.sativus L.).Its implications in future breeding programmes,in:A.K.Kaul,D.Combes(Eds.),Lathyrus and Lathyrism,Third World Medical Research Foundation,New York,USA 1986,pp.161-168.
[60]B.S.Dahiya,L.M.Jeswani,Estimation of genetic variance by full sib and half sib analysis in grass pea,Indian J.Agric.Sci. 44(1974)829-832.
[61]C.L.L.Gowda,A.K.Raul,Pulses in Bangladesh,Bangladesh Agricultural Research Institute and FAO,Dhaka,Bangladesh,1982.
[62]K.R.Tiwari,C.G.Campbell,Inheritance of neurotoxin(ODAP)content,flower and seed coat colour in grass pea(Lathyrus sativus L.),Euphytica 91(1996)195-203.
[63]R.B.Mehra,V.Kumari,G.K.Barat,D.B.Raju,K.Himabindu,Behavior of neurotoxin content in some crosses of grass pea(Lathyrus sativus L.),Lathyrus Lathyrism Newsl.2(1993)8.
[64]R.L.Pandey,P.Shrivastava,A.K.Geda,R.N.Sharma,Relative contribution of yield components and their relationship with neurotoxin content in grass pea(Lathyrus sativus L.),Ann.Agric.Res.21(2000)11-16.
[65]R.N.Sharma,O.P.Kashyap,M.W.Chitale,R.L.Pandey,Genetic analysis for seed attributes over the years in grass pea(Lathyrus sativus L.),Indian J.Genet.57(1997)154-157.
[66]P.Shrivastava,R.L.Pandey,Studies on Genetic Variability and Diversity in Grass Pea(Lathyrus sativus)(M.Sc Thesis)Indira Gandhi Agricultural University,Raipur,1966.
[67]C.D.Hanbury,K.H.M.Siddique,N.W.Galwey,P.S.Cocks,Genotype-environment interaction for seed yield and ODAP concentration of Lathyrus sativus L.and L.cicera L.In Mediterranean-type environments,Euphytica 110(1999)445-460.
[68]Y.S.Nerker,Mutation studies in Lathyrus sativus,Indian J. Genet.76(1976)223-229.
[69]D.Talukdar,S.C.Biswas,A.K.Biswas,An induced dwarf mutant of grass pea,Indian J.Genet.61(2001)383-384.
[70]D.Talukdar,Dwarf mutations in grass pea(Lathyrus sativus L.):origin,morphology,inheritance and linkage studies,J. Genet.88(2009)165-175.
[71]D.Talukdar,A.K.Biswas,An induced internode mutant in grass pea,in:R.K.Das,S.Chatterjee,G.C.Sadhukhan,G.K. Manna(Eds.),Perspectives in Cytology and Genetics,All India Congress of Cytology and Genetics,Kalyani,India,vol. 12,2006,pp.267-271.
[72]V.N.Waghmare,D.N.Waghmare,R.B.Mehra,An induced fascinated mutant in grass pea(Lathyrus sativus L.),Indian J. Genet.61(2001)155-157.
[73]D.Talukdar,S.C.Biswas,A.K.Biswas,Induced mutation in grass pea(Lathyrus sativus L.),in:G.K.Manna,S.C.Roy(Eds.),Perspectives in Cytology and Genetics,All India Congress of Cytology and Genetics,Kalyani,India,vol.10,2001,pp.481-484.
[74]D.Talukdar,Isolation,Characterization and Genetic Evaluation of Different Induced Mutant Lines Recovered in Grass Pea(Lathyrus sativus L.)(Ph.D.Thesis)University of Kalyani,Kalyani,India,2008.
[75]D.Talukdar,S.C.Biswas,A.K.Biswas,An induced flower colour mutant in grass pea(Lathyrus sativus L.),Indian J. Genet.62(2002)162.
[76]A.B.Prasad,A.K.Das,Morphological variants in Khesari,Indian J.Genet.40(1980)172-175.
[77]D.Talukdar,A.K.Biswas,Induced seed coat colour mutations and their inheritance in grass pea(Lathyrus sativus L.),Indian J.Genet.65(2005)135-136.
[78]D.Talukdar,A.K.Biswas,Characterization of an induced mutant and its inheritance in grass pea(Lathyrus sativus L.),Indian J.Genet.62(2002)355-356.
[79]S.C.Biswas,A.K.Biswas,Induced translocation heterozygosity and sterility in Lathyrus sativus L,Bangladesh J.Bot.26(1997)131-136.
[80]D.Talukdar,Reciprocal translocations in grass pea(Lathyrus sativus L.):pattern of transmission,detection of multiple interchanges and their independence,J.Hered.101(2009)169-176.
[81]D.Talukdar,T.Talukdar,Leaf rolling and stem fasciation in grass pea(Lathyrus sativus L.)mutant are mediated through glutathione-dependent cellular and metabolic changes and associated with a metabolic diversion through cysteine during phenotypic reversal,Biomed.Res.Int.(2014)479180.
[82]D.Talukdar,Flower and pod production,abortion,leaf injury,yield and seed neurotoxin levels in stable dwarf mutant lines of grass pea(Lathyrus sativus L.)differing in salt stress responses,Int.J.Curr.Res.2(2011)46-54.
[83]D.Talukdar,Isolation and characterization of NaCl-tolerant mutations in two important legumes,Clitoria ternatea L.and Lathyrus sativus L.:induced mutagenesis and selection by salt stress,J.Med.Plants Res.5(2011)3619-3628.
[84]D.Talukdar,An induced glutathione-deficient mutant in grasspea(LathyrussativusL.):modificationinplantmorphology, alteration in antioxidant activities and increased sensitivity to cadmium,Biorem.Biodivers.Bioavailab.6(2012)75-86.
[85]D.Talukdar,Arsenic exposure modifies Fusarium wilt tolerance in grass pea(Lathyrus sativus L.)genotypes through modulation of antioxidant defense response,J.Plant Sci. Mol.Breed.2(2013)4.
[86]D.Talukdar,Allozyme variations in leaf esterase and root peroxidase isozymes and linkage with dwarfing genes in induced dwarf mutants of grass pea(Lathyrus sativus L.),Int. J.Genet.Mol.Biol.2(2010)112-120.
[87]D.Talukdar,Ascorbate deficient semi-dwarf asfL1 mutant of Lathyrus sativus exhibits alterations in antioxidant defense,Biol.Plant.56(2012)675-682.
[88]D.Talukdar,Flavonoid-deficient mutants in grass pea(Lathyrus sativus L.):genetic control,linkage relationships,and mapping with aconitase and S-nitrosoglutathione reductase isozyme loci,Sci.World J.(2012)345983.
[89]V.G.Narsinghani,S.M.Kumar,The field reaction to powdery and downy mildew in Lathyrus genetic stock,J.Mycol.Plant Pathol.9(1979)252-253.
[90]M.S.Lal,I.Agrawal,M.W.Chitale,Genetic improvement of chickling vetch in Madhya Pradesh,India,in:A.K.Kaul,D. Combes(Eds.),Lathyrus and Lathyrism,Third World Medical Research Foundation,New York,USA 1985,pp.146-160.
[91]R.P.Mishra,S.R.Kotasthane,M.N.Kare,O.Gupta,S.P.Tiwari,Screening of germplasm collections of Lathyrus spp.against Cercospora pisi sativae f.sp.lathyri sativae,Indian J.Mycol. Plant Pathol.163(1986)302.
[92]M.C.Vaz Patto,D.Rubiales,Identification and characterization of partial resistance to rust in a germplasm collection of Lathyrus sativus,Plant Breed.128(2009)495-500.
[93]M.Fernández-Aparicio,F.Flores,D.Rubiales,Field response of Lathyrus cicera germplasm to crenate broomrape(Orobanche crenata),Field Crop Res.113(2009)321-327.
[94]H.K.Jain,N.Somayajulu,G.K.Barat,Final Technical Report on Investigation in Lathyrus sativus,Indian Agricultural Research Institute,New Delhi,India,1994.
[95]K.L.Sethi,R.B.Mehra,B.M.Lal,Studies on variability and correlation between toxin content and seed weight in Lathyrus sativus,Sci.Cult.47(1981)263-264.
[96]I.M.Santha,K.Ali,S.L.Mehta,Performance of low ODAP somaclones of Lathyrus sativus,in:P.N.Mathur,V. Ramanatha Rao,R.K.Arora(Eds.),Lathyrus Genetic Resources Network:Proceedings of a IPGRI-ICARDA-ICAR Regional Working Group Meeting,December 8-10,1997,National Bureau of Plant Genetic Resources,New Delhi 1998,pp.74-78(IPGRI Office for South Asia,New Delhi,India).
[97]S.Kumar,M.Imtiaz,S.Gupta,A.Pratap,Distant hybridization and alien gene introgression,in:A.Pratap,J.Kumar(Eds.),Biology and Breeding of Food Legumes,CAB International,Oxfordshire,United Kingdom 2011,pp.81-110.
[98]V.A.Aletor,A.M.Abd-El-Moneim,A.V.Goodchild,Evaluation of the seeds of selected lines of three Lathyrus spp.for b-N-oxalyl amino-L-alanine(BOAA),tannins,trypsin inhibitor activity and certain in vitro characteristics,J.Sci.Food Agric.65(1994)143-151.
[99]K.H.M.Siddique,S.P.Loss,S.P.Herwig,J.M.Wilson,Growth,yield and neurotoxin(ODAP)concentration of three Lathyrus species in Mediterranean type environments of Western Australia,Aust.J.Exp.Agric.36(1996)209-218.
[100]C.D.Hanbury,K.H.M.Siddique,N.W.Galwey,P.S.Cocks,Genotype environment interaction for seed yield and ODAP concentration of Lathyrus sativus L.and L.cicera L.in Mediterranean-type environments,Euphytica 110(1999)45-60.
[101]M.Zhou,R.K.Arora,Conservation and use of underutilized cropsinAsia,in:R.K.Arora,P.N.Mathur,K.W.Riley,Y.Adham(Eds.),Lathyrus Genetic Resources in Asia,International Plant Genetic Resources Institute,Rome,Italy 1996,pp.91-96.
[102]J.C.Sillero,J.I.Cubero,M.Fernández-Aparicio,D.Rubiales,Search for resistance to crenate broomrape(Orobanche crenata)in Lathyrus,Lathyrus Lathyrism Newsl.4(2005)7-9.
[103]M.Fernández-Aparicio,F.Flores,D.Rubiales,Field response of Lathyrus cicera germplasm to crenate broomrape(Orobanche crenata),Field Crop Res.113(2009)187-358.
[104]M.Fernández-Aparicio,D.Rubiales,Characterisation of resistance to crenate broomrape(Orobanche crenata Forsk.)Lathyrus cicera L,Euphytica 173(2010)77-84.
[105]L.D.Robertson,A.M.Abd-El-Moneim,Status of Lathyrus germplasm held at ICARDA and its use in breeding programs,in:P.N.Mathur,V.R.Rao,R.K.Arora(Eds.),Lathyrus Genetic Resources Network:Proceedings of a IPGRI-ICARDA-ICAR Regional Working Group Meeting,December 8-10,1997,National Bureau of Plant Genetic Resources,New Delhi 1998,pp.30-41(IPGRI Office for South Asia,New Delhi,India).
[106]A.J.S.Davies,Successful crossing in the genus Lathyrus through stylar amputation,Nature 180(1957)612.
[107]H.I.T.Khawaja,A new inter-specific Lathyrus hybrid to introduce the yellow flower color into sweat pea,Euphytica 37(1988)69-75.
[108]A.G.Yunus,Biosystematics of Lathyrus Section Lathyrus with Special Reference to the Grasspea,L.sativus L(Ph.D.Dissertation)University of Birmingham,United Kingdom,1990.
[109]K.Yamamoto,T.Fujiware,L.Blumenreich,Isozymic variation and interspecific crossability in annual species of the genus Lathyrus L,in:A.K.Kaul,D.Combes(Eds.),Lathyrus and Lathyrism,Third World Medical Research Foundation,New York 1989,pp.118-121.
[110]K.R.W.Hammett,B.G.Murray,K.R.Markham,I.C.Hallett,Interspecific hybridization between Lathyrus odoratus and L. belinensis,Int.J.Plant Sci.155(1994)763-771.
[111]K.R.W.Hammett,B.G.Murray,K.R.Markham,I.C.Hallett,I. Osterloh,New interspecific hybrids in Lathyrus(Leguminosae):Lathyrus annuus×L.hierosolymitanus,Bot.J. Linn.Soc.122(1996)89-101.
[112]A.J.S.Davies,A Cytogenetic Study in the Genus Lathyrus(Ph.D.Thesis)University of Manchester,United Kindom,1958.
[113]D.A.Trankovskij,Interspecific hybridization in the genus Lathyrus,Bull.Mosc.Nat.Biol.Ser.67(1962)140-141.
[114]J.P.Kearney,Wild Lathyrus Species as Genetic Resources for Improvement of Grasspea(L.sativus)(Ph.D.Dissertation)University of Southampton,United Kingdom,1993.
[115]M.Marsden-Jones,Hybrids of Lathyrus,J.R.Hortic.Soc.45(1919)92-93.
[116]S.J.Ochatt,P.Durieu,L.Jacas,C.Pontecaille,Protoplast,cell and tissue cultures for the biotechnological breeding of grasspea(Lathyrus sativus L.),Lathyrus Lathyrism Newsl.2(2001)35-38.
[117]A.Bohra,U.C.Jha,P.B.Kavi Kishor,S.Pandey,N.P.Singh,Genomics and molecular breeding in lesser explored pulse crops:current trends and future opportunities,Biotechnol. Adv.32(2014)1410-1428.
[118]D.Barik,L.Acharya,A.Mukherjee,P.Chand,Analysis of genetic diversity among selected grass pea(Lathyrus sativus L.)genotypes using RAPD markers,Z.Naturforsch.62(2007)869-874.
[119]N.Chtourou-Ghorbel,B.Lauga,D.Combes,M.Marrakchi,Comparative genetic diversity studies in the genus Lathyrus using RFLP and RAPD markers,Lathyrus Lathyrism Newsl.2(2001)62-68.
[120]S.Tavoletti,L.Iommarini,Molecular marker analysis of genetic variation characterizing a grass pea(Lathyrus sativus)collection from central Italy,Plant Breed.126(2007)607-611.
[121]H.Nosrati,M.A.Hosseinpour-Feizi,M.Nikniazi,A. Razban-Haghighi,Genetic variation among different accessions of Lathyrus sativus(Fabaceae)revealed by RAPDs,Bot.Serbica 36(2012)41-47.
[122]G.J.Kenicer,T.Kajita,R.T.Pennington,J.Murata,Systematics and biogeography of Lathyrus based on internal transcribed spacer and cpDNA sequence data,Am.J.Bot.92(2005)1199-1209.
[123]L.Lioi,F.Sparvoli,G.Sonnante,G.Laghetti,F.Lupo,M. Zaccardelli,Characterization of Italian grass pea(Lathyrus sativus L.)germplasm using agronomic traits,biochemical and molecularmarkers,Genet.Resour.Crop.Evol.58(2011)425-437.
[124]A.Chandra,Use of EST database markers from Medicago truncatula in the transferability to other forage legumes,J. Environ.Biol.32(2011)347-354.
[125]L.Lioi,I.Galasso,Development of genomic simple sequence repeat markers from an enriched genomic library of grass pea(Lathyrus sativus L.),Plant Breed.132(2013)649-653.
[126]K.R.Soren,A.Yadav,G.Pandey,P.Gangwar,A.K.Parihar,A. Bohra,G.P.Dixit,S.Datta,N.P.Singh,EST-SSR analysis provides insights about genetic relatedness,population structure and gene flow in grass pea(Lathyrus sativus),Plant Breed.134(2015)338-344.
[127]E.Shiferaw,M.E.Pè,E.Porceddu,M.Ponnaiah,Exploring the genetic diversity of ethiopian grass pea(Lathyrus sativus L.)using EST-SSR markers,Mol.Breed.30(2012)789-797.
[128]X.L.Sun,T.Yang,J.P.Guan,Y.Ma,J.Y.Jiang,R.Cao,M. Burlyaeva,M.Vishnyakova,E.Semenova,S.Bulyntsev,X.X. Zong,Development of 161 novel EST-SSR markers from Lathyrus sativus(Fabaceae),Am.J.Bot.99(2012)379-390.
[129]M.V.Gutierrez,M.C.Vaz Patto,T.Huguet,J.I.Cubero,M.T. Moreno,A.M.Torres,Cross-species amplification of Medicago truncatula microsatellites across three major pulse crops,Theor.Appl.Genet.110(2005)1210-1217.
[130]M.Ponnaiah,E.Shiferaw,M.E.Pe,E.Porceddu,Development and application of EST-SSRs for diversity analysis in Ethiopian grass pea,Plant Genet.Resour.9(2011)276-280.
[131]N.F.Almeida,S.Trindade Leitão,C.Caminero,A.M.Torres,D.Rubiales,M.C.V.Patto,Transferability of molecular markers from major legumes to Lathyrus spp.for their application in mapping and diversity studies,Mol.Biol.Rep. 41(2014)269-283.
[132]M.A.Chowdhury,A.E.Slinkard,Linkage of random amplified polymorphic DNA,isozyme and morphological markers in grass pea(Lathyrus sativus),J.Agric.Sci.33(1999)389-395.
[133]B.Skiba,R.Ford,E.C.Pang,Construction of a linkage map based on a Lathyrus sativus backcross population and preliminary investigation of QTLs associated with resistance to ascochyta blight,Theor.Appl.Genet.109(2004)1726-1735.
[134]T.Yang,J.Y.Jiang,M.Burlyaeva,J.G.Hu,C.J.Coyne,S. Kumar,R.Redden,X.L.Sun,F.Wang,J.W.Chang,X.P.Hao,J.P.Guan,X.X.Zong,Large-scale microsatellite development in grass pea(Lathyrus sativus L.)an orphan legume of the arid areas,BMC Plant Biol.14(2014)65.
[135]F.Wang,T.Yang,M.Burlyaeva,L.Li,J.Y.Jiang,L.Fang,R. Redden,X.X.Zong,Genetic diversity of grass pea and its relative species revealed by SSR markers,PLoS One 10(2015),e0118542.
[136]N.F.Almeida,S.T.Leitão,N.Krezdorn,B.Rotter,P.Winter,D. Rubiales,M.C.V.Patto,Allelic diversity in the transcriptomes of contrasting rust-infected genotypes of Lathyrus sativus,a lasting resource for smart breeding,BMC Plant Biol.14(2014)376.
[137]N.F.Almeida,N.Krezdorn,B.Rotter,P.Winter,D.Rubiales,M.C.V.Patto,Lathyrus sativus transcriptome resistance response to Ascochyta lathyri investigated by deepSuperSAGE analysis,Front.Plant Sci.6(2015)178.
28 May 2016
in revised form 29 June 2016 Accepted 11 July 2016
.
E-mail address:ashoka.parihar@gmail.com(A.K.Parihar).
Peer review under responsibility of Crop Science Society of China and Institute of Crop Science,CAAS.
1These authors contributed equally to this work.
http://dx.doi.org/10.1016/j.cj.2016.06.008
2214-5141/©2016 Crop Science Society of China and Institute of Crop Science,CAAS.Production and hosting by Elsevier B.V.This is an open access article under the CC BY-NC-ND license(http://creativecommons.org/licenses/by-nc-nd/4.0/).
杂志排行
The Crop Journal的其它文章
- Development of a panel of unigene-derived polymorphic EST-SSR markers in lentil using public database information
- Characterization of chickpea germplasm conserved in the Indian National Genebank and development of a core set using qualitative and quantitative trait data
- Nutritional composition and antioxidant activity of twenty mung bean cultivars in China
- Genetics of seed flavonoid content and antioxidant activity in cowpea(Vigna unguiculata L.Walp.)
- Molecular cloning and characterization of a gene encoding the proline transporter protein in common bean(Phaseolus vulgaris L.)
- Large-scale evaluation of pea(Pisum sativum L.)germplasm for cold tolerance in the field during winter in Qingdao
