光照、糖和激素对花青素合成调控的综述
2016-10-17王海竹徐启江闫海芳
王海竹,徐启江,闫海芳
(1.东北林业大学 林木遗传育种国家重点实验室,黑龙江 哈尔滨 150040;2.东北林业大学 生命科学学院,黑龙江 哈尔滨 150040)
光照、糖和激素对花青素合成调控的综述
王海竹1,2,徐启江1,2,闫海芳2*
(1.东北林业大学 林木遗传育种国家重点实验室,黑龙江 哈尔滨 150040;2.东北林业大学 生命科学学院,黑龙江 哈尔滨 150040)
花青素是一类黄酮类化合物,存在于植物的叶片、花、果实和种子的表皮细胞的液泡中,花青素合成不仅受到系列结构基因和转录因子的调控,还受到光照、温度、糖、激素、pH、温度和氮等因素影响。综述了光照、糖和激素对花青素生物合成途径调控作用方面的研究进展。
糖;光照;激素;花青素合成
花青素是植物体内一类次生代谢物质,广泛存在于开花植物(被子植物)中,据初步统计,已发现有20种花青素,但可作为食品添加剂的仅有6种[1]。花青素无毒、无特殊气味,具有多种营养、药理和保健功能,在食品、化妆、医药方面有着巨大的应用潜力。
1 花青素的生物合成
花青素的合成是一个多酶途径[2],包括苯丙氨酸解氨酶(PAL)、肉桂-4-羟化酶(C4H)、4-香豆酸辅酶A连接酶(4CL)、查耳酮合酶(CHS)、查耳酮异构酶(CHI)、黄烷酮三羟化酶(F3H)、类黄酮3′-羟化酶(F3′H)、类黄酮3′,5′-羟化酶(F3′5′H)、二氢黄酮醇-4-还原酶(DFR)、花青素合成酶/无色花青素双加氧酶(ANS/LDOX)、类黄酮-3-O-葡萄糖基转移酶(UFGT)。PAL是此过程中的第一个酶,催化苯丙氨酸变成肉桂酸,而UFGT在最后一步使得花色苷转变成稳定的花青素[3]。
花青素合成途径结构基因的表达直接影响到花青素含量[4-5]。在自然条件下,与花青素合成相关且具有转录活性的转录因子能够在特定组织中上调结构基因的表达[6-7],转录因子包括R2R3-MYB转录因子、bHLH转录因子和WD40蛋白,这3种转录因子常常结合形成转录复合物调节结构基因的表达[8-9]。
花青素合成除了受结构基因表达水平影响外,还受光照、糖、激素、pH、温度、氮、磷等因素所支配。本文主要综述了光照、糖和激素对花青素合成的影响。
2 光照对花青素合成的调控
光作为最重要的环境因子之一,植物通过光受体可以感受光的强度、方向、光周期等,并通过信号转导途径调控自身的光形态建成、开花诱导、生物节律以及代谢产物的合成等多个生长发育过程[10]。
光信号通过不同的光受体作用于各种生理过程,目前发现的光受体有:吸收红光和远红光的光敏色素(PHYA、PHYB、PHYC、PHYD、PHYE)[11]、吸收蓝光和UV-A的隐花色素(CRY1、CRY2、CRY3)[12]和向光素(PHOT1、PHOT2)以及UV-B敏感光受体(UVR8)[11,13-15]。
光敏色素(phytochrome)能够调节植物的生长发育进程,例如种子萌发、下胚轴延长、开花等[16-18],此外COP1是一个指环结构的E3泛素连接酶,是光形态建成的负调控因子,在拟南芥中发现COP1与光敏色素、隐花色素和向光素存在蛋白之间相互作用[19-20]。而PHYA和PHYB相互作用能够抑制COP1的活性;COP1/SPA复合物还能够启动PHYA泛素化,从而削弱远红光信号传递途径[21]。
隐花色素(cryptochrome)是一种黄素蛋白受体,参与许多次级代谢产物的生物合成、植物发育进程和生物节律信号传递过程,推测可能存在2条CRY介导的光信号传导途径[22]。最近有研究表明,向光素在蓝光介导次级代谢产物生物合成过程具有重要作用,具体机制尚不清楚[23]。
光信号除了通过光受体影响植物发育进程,还可以利用信号转导因子调控启动子与转录子结合的强弱,直接或间接调节一些关键酶的合成,从而调节花青素合成途径中结构基因的表达,进而影响花青素的合成[24-25]。经过对葡萄、苹果、梨等植物研究发现很多与花青素合成相关的关键转录因子,R2R3-MYB作为主要调节因子通过激活或抑制合成途径中结构基因的表达来直接影响花青素的合成[26-28]。研究者发现一些R2R3-MYB转录因子也能够响应光信号,HY5是一个具有锌指结构的R2R3-MYB转录因子,也是COP1的靶基因,能够启动光形态建成[25,29-30]。在可见光条件下,COP1从细胞核中移出,HY5便能够稳定存在,与其他转录因子形成复合物启动花青素合成结构基因表达,产生花青素;在黑暗条件下,COP1存在于细胞核中,COP1/SPA复合物使得HY5泛素化,进而被降解,从而抑制花青素的合成,并且在cop1突变体中,明显发现花青素积累[31-32]。与在可见光下不同,UV-B照射下,COP1是一个正调控因子,并且UV-B光信号受体UVB8由二聚体变成单体,此时的单体会与COP1相互作用促进HY5的表达,从而增加花青素含量,但是有研究者也发现在葡萄中却没有这种响应机制[33-35],并推测可能存在响应UV-A的光受体,但目前仍不能确定[35-36]。
3 糖对花青素合成的调控
糖是作为常见调节因子,能够调节一些参与光合作用、糖代谢和花青素合成等的信号表达,其信号途径已经在细菌[37]等微生物中得到研究和应用。蔗糖作为外源物质可以调节葡萄果皮中花青素合成基因表达[38];拟南芥经外源蔗糖、葡萄糖和果糖处理12 h后,花青素苷合成结构基因CHS、CHI、F3H、F3′H、DFR和LDQZ的表达量均明显上调[39]。蔗糖不仅可以调控结构基因的表达,而且还可以特异性地调控调节基因的表达,但是葡萄糖和果糖对MYB75/PAP1的表达无影响[39]。此外,还发现蔗糖对花青素合成结构基因的影响要超过正调控因子GL3、TT8和PAP1等,超过了负调控因子MYBL2的调控能力[40]。
拟南芥中有9种蔗糖转运蛋白(Suc transporters,SUCs),光照、温度和糖等因素会影响SUCs的表达[40]。不同种类的糖可以对SUC1基因的表达和花青素的积累产生不同的效果,蔗糖和麦芽糖等一些可代谢类双糖能够调节花青素的形成,而蔗糖受体基因SUC1的表达主要受单糖的诱导和调节,如葡萄糖和果糖,甘露糖却不能产生同样的效果。蔗糖和麦芽糖等二糖或葡萄糖和果糖的分解产物都能够激活糖信号途径,进而调节花青素合成,并且花青素合成被蔗糖和麦芽糖的二糖代谢优先诱导,而葡萄糖和果糖等单糖能有效诱导SUC1表达[40]。但是蔗糖转运蛋白SUC1在植物根中表达[41],花青素积累则在叶表皮细胞[40,42],因此在根中表达的SUC1如何参与叶中花青素合成仍需要进一步探索。
4 激素对花青素合成调控
4.1生长素对花青素合成的调控
生长素作为植物生理过程中重要的激素,能够调控植物的器官发生和形态建成[43-44]、黄酮类积累[45]等。生物体内生长素主要以天然的吲哚-3-乙酸(Indole-3-acetic acaid,IAA)的形式存在[46]。用IAA处理拟南芥幼苗,发现花青素合成途径结构基因CHS、CHI、F3′H的表达量上调,相关转录因子TTG1、PAP1和MYB12的表达量也增加,说明生长素对花青素合成具有一定的促进作用[45]。
在生长素信号途径中,生长素/吲哚乙酸蛋白基因家族(AUX/IAAs),是一类转录抑制因子,能够被生长素响应因子蛋白家族(ARFs)降解,在此过程中ARFs作为正调控因子增强靶基因表达,从而影响植物生长与发育[47];运输抑制剂响应蛋白1(TIP1)目前发现的一种真正意义上的生长素受体,是一个E3泛素连接酶,能够降解转录抑制因子,从而诱导基因表达[48]。在突变体tip1中,发现转录因子TTG1、PAP1和MYB12表达量减少相当明显,表明TIP1在生长素影响花青素合成途径具有重要作用[45]。
4.2细胞分裂素对花青素合成的调控
细胞分裂素(cytokinin,CK)对花青素合成的促进作用已经在很多植物中得到验证,如胡萝卜(DaucuscarotaL.var.carota)、玫瑰(Rosaceae)和油菜(BrassicacampestrisL.)等[49]。用细胞分裂素同样处理长在MS培养基上的拟南芥幼苗,在黑暗条件下没有花青素积累,而在光照条件下可以明显观察到有花青素的合成,由此说明细胞分裂素促进花青素合成需要光照的存在[50]。
细胞分裂素信号途径参与花青素合成的细胞分裂素受体有组蛋白激酶(AHK2、AHK3和AHK4)和B型响应调节因子(ARR1、ARR10和ARR12)[51]。组蛋白激酶是细胞分裂素信号途径正调控因子[52-54],用糖和细胞分裂素处理双重突变体ahk2/3和ahk3/4后明显发现花青素积累减少了[51]。B型响应调节因子(ARRs)能够激活上游A型响应调节因子ARR基因表达[55-56]。在拟南芥中发现的11种B型响应调节因子(ARRs),其中有7种ARRs与细胞分裂素信号途径有关[57]。研究者发现在单突变体arr1、arr10和arr12,双突变体arr1/10、arr1/12和arr10/12,三重突变体arr1/10/12中,蔗糖诱导花青素合成途径中花青素积累较少,此研究表明:细胞分裂素通过光电子传递信号途径转录激活正调控因子PAP1、(E)GL3和TT8,抑制负调控因子MYBL2转录水平,从而参与蔗糖诱导花青素积累[51,58]。
4.3脱落酸对花青素合成的调控
脱落酸(abscisic acid,ABA)是以异戊二烯为基本单位的倍半萜羧酸。脱落酸喷洒处理葡萄能够促进花青素的合成积累[59-60]。通过对拟南芥的研究发现,ABA单独处理植株,只影响PAP2和ATT表达;当有糖存在时,ABA处理植株发现大部分与花青素合成相关基因的表达都呈现上调趋势包括PAP1,表明ABA促进花青素合成可能需要糖的存在[61-62]。也有研究者认为ABA对花青素合成的影响并严格依赖于糖的存在。
在玉米ABA不敏感突变体vp1研究中发现一个与PAP1和PAP2功能相近的MYB正调控转录因子C1基因不表达,并且该基因启动子活性受ABA调节,间接证明ABA与花青素合成有关[63]。ABA信号与一些延伸因子相关,这些延伸因子的突变体植株,部分与花青素合成相关基因的表达呈现下调趋势,由于延伸因子是在mRNA翻译时促进多肽链延伸的蛋白质因子,因而研究者猜测其作用机制可能是直接作用于MYBL2基因转录延伸[64],进而参与调节花青素生物合成。但是ABA使得花青素积累究竟是通过增加ABA含量还是影响ABA信号转导仍需要进一步研究。
4.4赤霉素对花青素合成的调控
赤霉素(gibberellin,GA)是一类四环二萜类化合物,是植物六大激素之一,在植物的胚胎发育、种子休眠、果实成熟及逆境胁迫等许多方面存在广泛的生理效应,也是花青素合成途径中负调控因子[65]。大多数GAs都是生物活性物质的前体或是失活的代谢物,只有少数(GA1、GA3、GA4和GA7)具有生物学活性[66]。
利用缺乏赤霉素应答的拟南芥突变植株ga1,发现该突变体无法完成催化赤霉素合成途径中牻牛儿基焦磷酸向古巴焦磷酸转变的合成,直接导致GA前体无法生成,从而使得具有活性的内源赤霉素的含量较低,抑制作用降低,进而触发了参与花青素合成途径基因PAP1等基因表达上调,因此植株花青素含量增加[61]。还发现GA3能够抑制花青素合成途径中结构基因DFR和转录因子PAP1和PAP2表达水平[61]。
赤霉素抑制花青素合成依赖于赤霉素信号途径中负调控因子DELLA蛋白,该蛋白在拟南芥中有5种,即GAI、RGA、RGLA1、RGL2和RGL3[67-68],作用于GA受体,但并不影响植物体内赤霉素含量[69]。DELLA位于细胞核内,属于GRAS转录调节因子家族[70],能有效降解是GA发挥正常生理功能的标志,GA结合其可溶性受体GID1引起GID1构象变化,该构象下的GID4-GA能够与DELLAs结合并形成复合物,随后此复合物结合到SCF复合物上,依赖于SCFSLY1E3泛素连接酶,经泛素-蛋白酶体途径降解[71-72]。通过观察gai突变体表型发现突变体植株明显呈现深绿色,说明DELLA在GA抑制花青素合成中发挥重要作用[73]。
4.5乙烯对花青素合成的调控
乙烯对花青素合成抑制作用已经在很多实验中被证实,例如在黑暗条件下用乙烯处理甘蓝,其叶片颜色由红色变成白色[74-75]。转基因烟草中转入的乙烯受体基因证明其在乙烯抑制花青素合成中有重要作用。在烟草花瓣中过表达突变的乙烯受体基因(ETR1H69A)[76],花青素含量会增加。
在拟南芥的内质网上发现能够感知乙烯信号的受体,如ETR1、ETR2、ERS1、ERS2和EIN4等,由ETRs和ERSs等编码的与下游类似于Raf的蛋白激酶CTR1协同负调控乙烯反应;EIN2及其下游的EIN3/EILs位于CTR1下游,正调控乙烯反应。在突变体etr1-1、ein2-1、ein3和eil1拟南芥植株中花青素含量明显增加,同时这些受体在功能上有重叠,如在ein3和eil1双重突变体中花青素含量显著增加[58]。因此,乙烯诱导花青素合成途径的抑制作用可能涉及乙烯三重反应[40]。
乙烯抑制花青素积累常常通过在转录水平下调正调控转录因子bHLHs(GL3、EGL3和TT8)和MYBs(PAP1和PAP2)以及上调负调控转录因子MYBL2[58]。当有乙烯信号突变体存在时,抑制乙烯启动MYBL2表达,进而无法形成MYBL2-bHLH-WD40(M(L2)BW)转录复合物,或者即使M(L2)BW转录复合物能够形成,还存在高浓度有活性的MYB-bHLH-WD40(MBW)复合物,在这种情况下MBW复合物占主导位置,因此花青素含量增加。但是乙烯信号途径中关键的转录因子是否直接调节M(L2)BW转录复合物仍需要进一步研究。
4.6茉莉酸对花青素合成的调控
茉莉酸家族(JAs)包括茉莉酸、茉莉酸前体环戊烷和环戊烯酮,能够促进花青素积累[61]。他们可以响应许多生物和非生物胁迫[77-79]。对于拟南芥幼苗,仅仅用茉莉酸处理并不能影响花青素生物合成途径和转录因子PAP1和PAP2表达。当茉莉酸和糖同时存在时,茉莉酸能够增强合成途径中CHI和其下游基因以及PAP1和PAP2的表达水平,这种协同影响常常表现在mRNA水平和相关的花青素积累上。因此糖的存在是茉莉酸发挥作用的前提。
研究者通过对拟南芥茉莉酸信号途径冠菌素不敏感突变体coi1和茉莉酸不敏感突变体jar1的研究发现[80-81]:拟南芥对茉莉酸的响应需要COI1存在,因此冠菌素不敏感突变体coi1不能表达经茉莉酸诱导的基因[80,82-83]。Kim和Devoto等利用糖存在的情况下用茉莉酸处理拟南芥coi1突变体,结果显示并未检测到参与花青素合成基因的表达[59,84-85]。2008年Loreti等选用coi1同源突变体coi1-1植株,用其叶条作为实验材料,发现用糖处理与用糖和茉莉酸共同处理的coi1-1植株叶条的DFR基因表达都被正调控,仅仅用茉莉酸处理的coi1-1植株叶条只是在光合作用组织中含糖量增加,但是与野生型相比,不论是用糖处理,还是用糖与茉莉酸共同处理的植株叶条相比DFR基因的表达还是减少,此研究结果表明,在coi1-1突变体植株中不仅仅茉莉酸几乎发挥作用,而且糖对花青素合成途径正调控作用也受到一定程度的影响[61]。综上所述,COI1是茉莉酸介导的花青素合成途径所必需的。
研究者发现了一类JAZ蛋白家族,该蛋白家族是茉莉酸信号途径中关键的调控蛋白[86],是SCFCOI1复合物的作用底物,是茉莉酸信号途径的负调控因子[87-88]。通过蛋白质之间相互作用研究发现JAZs可以直接与MBW复合物中bHLHs(TT8、GL3和EGL3)和R2R3-MYBs转录因子(PAP1和GL1)相互作用[89],进而影响花青素合成。近期在茉莉酸信号途径中发现:COI1在茉莉酸途径中也具有重要作用,COI1可以招募JAZs到SCFCOI1复合物上使其泛素化,随后被蛋白酶体降解,使得JAZs释放MBW复合物,从而激活茉莉酸诱导的花青素合成。
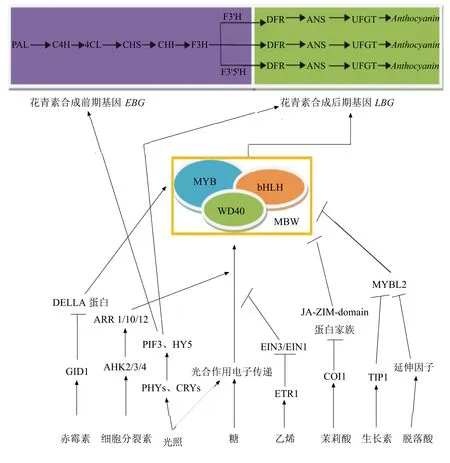
注:PAL为苯丙氨酸解氨酶;C4H为肉桂酸经化酶;4CL为香豆酰-CoA连接酶;CHS为查尔酮合成酶;CHI为查尔酮异构酶;F3H为黄烷酮-3-羟基化酶;F3′H为类黄酮-3′-羟基化酶;F3′5′H为类黄酮-3′, 5′-羟基化酶;DFR为二氢黄酮醇还原酶;ANS为花青素合成酶;UFGT为UDP-葡萄糖类黄酮-3-葡萄糖转移酶;Anthocyanin为花青素;→表示促进作用;┴表示抑制作用。
图1光照、糖和激素调控花青素合成示意图
5 结语
花青素合成受到光照、糖和激素的信号途径的调控如图1,除此之外也可能与Ca2+信号途径有关,但具体的机制并不清楚。很多研究已经表明光照是激素调节的花青素合成过程关键因子,而且糖和激素之间具体的相互作用也被阐明[58]。接下来可能要着眼于研究在UVR8、糖或激素等其他刺激的影响下,相关转录因子转录和转录后调节,并且不同激素之间相互作用影响花青素合成也需要考虑。未来可以通过在蛋白质和基因组水平更深层次明确花青素合成调节。
[1] Glover B J, Martin C. Anthocyanins[J]. Current Biology, 2012, 22(5): R147-R150.
[2] Zhang Y, Hu Z, Chu G, et al. Anthocyanin accumulation and molecular analysis of anthocyanin biosynthesis-associated genes in eggplant (SolanummelongenaL.)[J]. Journal of Agricultural and Food Chemistry, 2014, 62(13): 2906-2912.
[3] Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology[J]. Plant Physiology, 2001, 126(2): 485-493.
[4] Crifò T, Petrone G, Lo Cicero L, et al. Short cold storage enhances the anthocyanin contents and level of transcripts related to their biosynthesis in blood oranges[J]. Journal of Agricultural and Food Chemistry, 2011, 60(1): 476-481.
[5] Yuan Y, Chiu L W, Li L. Transcriptional regulation of anthocyanin biosynthesis in red cabbage[J]. Planta, 2009, 230(6): 1141-1153.
[6] Butelli E, Licciardello C, Zhang Y, et al. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges[J]. Plant Cell, 2012, 24(3): 1242-1255.
[7] Chiu L W, Zhou X, Burke S, et al. The purple cauliflower arises from activation of aMYBtranscription factor[J]. Plant Physiology, 2010, 154(3): 1470-1480.
[8] Albert N W, Lewis D H, Zhang H, et al. Members of anR2R3-MYBtranscription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning[J]. Plant J, 2011, 65(5): 771-784.
[9] Spelt C, Quattrocchio F, Mol J N, et al. anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes[J]. Plant Cell, 2000, 12(9): 1619-1631.
[10] Batschauer A. Photoreceptors of higher plants[J]. Planta, 1998, 206(4): 479-492.
[11] Nagatani A. Phytochrome: structural basis for its functions[J]. Curr Opin Plant Biol, 2010, 13(5): 565-570.
[12] Chaves I, Pokorny R, Byrdin M, et al. The cryptochromes: blue light photoreceptors in plants and animals[J]. Annu Rev Plant Biol, 2011, 62(1): 335-364.
[13] Rizzini L, Favory J J, Cloix C, et al. Perception of UV-B by theArabidopsisUVR8 protein[J]. Science, 2011, 332(6025): 103-106.
[14] Casal J J. Photoreceptor signaling networks in plant responses to shade[J]. Annu Rev Plant Biol, 2013, 64(1): 403-427.
[15] Heijde M, Ulm R. UV-B photoreceptor-mediated signaling in plants[J]. Trends Plant Sci, 2012, 17(4): 230-237.
[16] Rockwell N C, Su Y-S, Lagarias J C. Phytochome structure and signaling mechanisms[J]. Annu Rev Plant Biol, 2006, 57(57): 837-858.
[17] Khanna R, Huq E, Kikis E A, et al. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors[J]. The Plant Cell, 2004, 16(11): 3033-3044.
[18] Leivar P, Quail P H.PIFs: pivotal components in a cellular signaling hub[J]. Trends Plant Sci, 2011, 16(1): 19-28.
[19] Jang I C, Henriques R, Seo H S, et al. ArabidopsisPHYTOCHROMEINTERACTINGFACTORproteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus[J]. The Plant Cell, 2010, 22(7): 2370-2383.
[20] Li J G, Li G, Wang H Y, et al. Phytochrome signaling mechanisms[J]. Arabidopsis Book, 2004, 3(3): e74.
[21] Zuo Z, Liu H, Liu B, et al. Blue light-dependent interaction ofCRY2 withSPA1 regulatesCOP1 activity and floral initiation inArabidopsis[J]. Curr Biol, 2011, 21(10): 841-847.
[22] Liu H, Yu X, Li K, et al. PhotoexcitedCRY2 interacts withCIB1 to regulate transcription and floral initiation inArabidopsis[J]. Science, 2008, 322(5907): 1535-1539.
[23] Kadomura-Ishikawa Y, Miyawaki K, Noji S, et al. Phototropin 2 is involved in blue light-induced anthocyanin accumulation inFragariaxananassafruits[J]. J Plant Res, 2013, 126(6): 847-857.
[24] Zoratti L, Karppinen K, Escobar A L, et al. Light-controlled flavonoid biosynthesis in fruits[J]. Frontiers in Plant Science, 2014, 5(5): 534.
[25] Lee J, He K, Stolc V, et al. Analysis of transcription factorHY5 genomic binding sites revealed its hierarchical role in light regulation of development[J]. The Plant Cell, 2007, 19(3): 731-749.
[26] Petroni K, Tonelli C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs[J]. Plant Sci, 2011, 181(3): 219-229.
[27] Czemmel S, Heppel S C, Bogs J.R2R3MYBtranscription factors: key regulators of the flavonoid biosynthetic pathway in grapevine[J]. Protoplasma, 2012, 249(2): 109-118.
[28] Jaakola L. New insights into the regulation of anthocyanin biosynthesis in fruits[J]. Trends Plant Sci, 2013, 18(9): 477-483.
[29] Stracke R, Favory J J, Gruber H, et al. TheArabidopsisbZIPtranscription factorHY5 regulates expression of thePFG1/MYB12 gene in response to light and ultraviolet-B radiation[J]. Plant, Cell Environ, 2010, 33(1): 88-103.
[30] Maier A, Schrader A, Kokkelink L, et al. Light and the E3 ubiquitin ligaseCOP1/SPAcontrol the protein stability of theMYBtranscription factorsPAP1 andPAP2 involved in anthocyanin accumulation in Arabidopsis[J]. The Plant Journal, 2013, 74(4): 638-651.
[31] Vierstra R D. The ubiquitin-26S proteasome system at the nexus of plant biology[J]. Nature Reviews Molecular Cell Biology, 2009, 10(6): 385-397.
[32] Lau O S, Deng X W. The photomorphogenic repressorsCOP1 andDET1: 20 years later[J]. Trends Plant Sci, 2012, 17(10): 584-593.
[33] Liu L, Gregan S, Winefield C, et al. From UVR8 to flavonol synthase: UV-B-induced gene expression in Sauvignon blanc grape berry[J]. Plant, Cell Environ, 2015, 38(5): 905-919.
[34] Peng T, Saito T, Honda C, et al. Screening of UV-B-induced genes from apple peels bySSH: possible involvement ofMdCOP1-mediated signaling cascade genes in anthocyanin accumulation[J]. Physiol Plant, 2013, 148(3): 432-444.
[35] Guo J, Wang M H. Ultraviolet A-specific induction of anthocyanin biosynthesis and PAL expression in tomato (SolanumlycopersicumL.)[J]. Plant growth regulation, 2010, 62(1): 1-8.
[36] Wang Y, Zhou B, Sun M, et al. UV-A light induces anthocyanin biosynthesis in a manner distinct from synergistic blue+ UV-B light and UV-A/blue light responses in different parts of the hypocotyls in turnip seedlings[J]. Plant and Cell Physiology, 2012, 53(8): 1470-1480.
[37] Rolland F, Baena-Gonzalez E, Sheen J. Sugar sensing and signaling in plants: conserved and novel mechanisms[J]. Annu Rev Plant Biol, 2006, 57(1): 675-709.
[38] Boss P K, Davies C, Robinson S P. Expression of anthocyanin biosynthesis pathway genes in red and white grapes[J]. Plant Mol Biol, 1996, 32(3): 565-569.
[39] Solfanelli C, Poggi A, Loreti E, et al. Sucrose-specific induction of the anthocyanin biosynthetic pathway inArabidopsis[J]. Plant Physiology, 2006, 140(2): 637-646.
[40] Jeong S W, Das P K, Jeoung S C, et al. Ethylene suppression of sugar-induced anthocyanin pigmentation inArabidopsis[J]. Plant Physiology, 2010,154(3): 1514-1531.
[41] Sivitz A B, Reinders A, Ward J M. Arabidopsis sucrose transporterAtSUC1 is important for pollen germination and sucrose-induced anthocyanin accumulation[J]. Plant Physiology, 2008, 147(1): 92-100.
[42] Kubo H, Peeters A J, Aarts M G, et al.ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis[J]. Plant Cell, 1999, 11(7): 1217-1226.
[43] Bohn-Courseau I. Auxin: a major regulator of organogenesis[J]. C R Biol, 2010, 333(4): 290-296.
[44] Reinhardt D. Vascular patterning: more than just auxin?[J]. Curr Biol, 2003, 13(12): R485-R487.
[45] Lewis D R, Ramirez M V, Miller N D, et al. Auxin and ethylene induce flavonol accumulation through distinct transcriptional networks[J]. Plant Physiology, 2011, 156(1): 144-164.
[46] Woodward A W, Bartel B. Auxin: regulation, action, and interaction[J]. Ann Bot, 2005, 95(5): 707-735.
[47] Shin R, Burch A Y, Huppert K A, et al. TheArabidopsistranscription factorMYB77 modulates auxin signal transduction[J]. The Plant Cell, 2007, 19(8): 2440-2453.
[48] Kepinski S, Leyser O. TheArabidopsisF-box proteinTIR1 is an auxin receptor[J]. Nature, 2005, 435(7041): 446-451.
[49] Nakamura N, Nakamae H, Maekawa S. Effects of light and kinetin on anthocyanin accumulation in the petals ofRosahybrida,Hortcv.Ehigasa[J]. Zeitschrift für Pflanzenphysiologie, 1980, 98(3): 263-270.
[50] Wade H K, Sohal A K, Jenkins G I.ArabidopsisICX1 is a negative regulator of several pathways regulating flavonoid biosynthesis genes[J]. Plant Physiology, 2003, 131(2): 707-715.
[51] Das P K, Shin D H, Choi S B, et al. Cytokinins enhance sugar-induced anthocyanin biosynthesis inArabidopsis[J]. Molecules and cells, 2012, 34(1): 93-101.
[52] Nishimura C, Ohashi Y, Sato S, et al. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth inArabidopsis[J]. Plant Cell, 2004, 16(6): 1365-1377.
[53] Riefler M, Novak O, Strnad M, et al.Arabidopsiscytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism[J]. Plant Cell, 2006, 18(1): 40-54.
[54] Tran L S P, Urao T, Qin F, et al. Functional analysis ofAHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress inArabidopsis[J]. Proceedings of the National Academy of Sciences, 2007, 104(51): 20623-20628.
[55] Argyros R D, Mathews D E, Chiang Y H, et al. Type B response regulators ofArabidopsisplay key roles in cytokinin signaling and plant development[J]. Plant Cell, 2008, 20(8): 2102-2118.
[56] Ishida K, Yamashino T, Yokoyama A, et al. Three type-B response regulators,ARR1,ARR10 andARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle ofArabidopsisthaliana[J]. Plant and Cell Physiology, 2008, 49(1): 47-57.
[57] Mason M G, Mathews D E, Argyros D A, et al. Multiple type-B response regulators mediate cytokinin signal transduction inArabidopsis[J]. Plant Cell, 2005, 17(11): 3007-3018.
[58] Das P K, Shin D H, Choi S B, et al. Sugar-hormone cross-talk in anthocyanin biosynthesis[J]. Molecules and cells, 2012, 34(6): 501-507.
[59] Kim J S, Lee B H, Kim S H, et al. Responses to environmental and chemical signals for anthocyanin biosynthesis in non-chlorophyllous corn (ZeamaysL.) leaf[J]. Journal of Plant Biology, 2006, 49(1): 16-25.
[60] Mori K, Sato H, Goto-Yamamoto N, et al. Effects of abscisic acid treatment and night temperatures on anthocyanin composition in Pinot noir grapes[J]. Progress in Neuro-Psychopharmacology and Biology, 2005, 29(2): 351-353.
[61] Loreti E, Povero G, Novi G, et al. Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes inArabidopsis[J]. New Phytol, 2008, 179(4): 1004-1016.
[62] Tonelli C, Cominelli E, Allegra D, et al. Plant tolerance to drought and salinity: modulation of transcription factors[C]//Proceedings of the 18th international conference onArabidopsisresearch. 2007.
[63] McCarty D R, Carson C B, Stinard P S, et al. Molecular analysis of viviparous-1: an abscisic acid-insensitive mutant of maize[J]. Plant Cell, 1989, 1(5): 523-532.
[64] Zhou G, Qi J, Ren N, et al. SilencingOsHI-LOXmakes rice more susceptible to chewing herbivores, but enhances resistance to a phloem feeder[J]. Plant J, 2009, 60(4): 638-648.
[65] Yamaguchi S. Gibberellin metabolism and its regulation[J]. Annu Rev Plant Biol, 2008, 59: 225-251.
[66] Peter H, Stephen G T. Gibberellin biosynthesis and its regulation[J]. Biochem J, 2012, 444(1): 11-25.
[67] Jiang C, Fu X. GA action: turning on de-DELLArepressing signaling[J]. Curr Opin Plant Biol, 2007, 10(5): 461-465.
[68] Wild M, Davière J-M, Cheminant S, et al. TheArabidopsisDELLARGA-LIKE3 is a direct target ofMYC2 and modulates jasmonate signaling responses[J]. The Plant Cell, 2012, 24(8): 3307-3319.
[69] Davière J M, Achard P. Gibberellin signaling in plants[J]. Development, 2013, 140(6): 1147-1151.
[70] Bolle C. The role ofGRASproteins in plant signal transduction and development[J]. Planta, 2004, 218(5): 683-692.
[71] Sun T P, Gubler F. Molecular mechanism of gibberellin signaling in plants[J]. Annu Rev Plant Biol, 2004, 55: 197-223.
[72] Ariizumi T, Lawrence P K, Steber C M. The role of two F-box proteins,SLEEPY1 andSNEEZY, in Arabidopsis gibberellin signaling[J]. Plant Physiology, 2011, 155(2): 765-775.
[73] Jiang C, Gao X, Liao L, et al. Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis[J]. Plant Physiology, 2007, 145(4): 1460-1470.
[74] Craker L, Wetherbee P J. Ethylene, light, and anthocyanin synthesis[J]. Plant physiology, 1973, 51(3): 436-438.
[75] Kang B G, Burg S P. Role of ethylene in phytochrome-induced anthocyanin synthesis[J]. Planta, 1973, 110(3): 227-235.
[76] Takada K, Ishimaru K, Minamisawa K, et al. Expression of a mutated melon ethylene receptor geneCm-ETR1/H69Aaffects stamen development inNicotianatabacum[J]. Plant science, 2005, 169(5): 935-942.
[77] Balbi V, Devoto A. Jasmonate signalling network inArabidopsisthaliana: crucial regulatory nodes and new physiological scenarios[J]. New Phytol, 2008, 177(2): 301-318.
[79] Turner J G, Ellis C, Devoto A. The jasmonate signal pathway[J]. Plant Cell, 2002, 14(suppl 1): S153-S164.
[80] Feys B J, Benedetti C E, Penfold C N, et al.Arabidopsismutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen[J]. Plant Cell, 1994, 6(5): 751-759.
[81] Staswick P E, Tiryaki I, Rowe M L. Jasmonate response locusJAR1 and several relatedArabidopsisgenes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation[J]. The Plant Cell, 2002, 14(6): 1405-1415.
[82] Benedetti C E, Xie D, Turner J G.COI1-dependent expression of an Arabidopsis vegetative storage protein in flowers and siliques and in response to coronatine or methyl jasmonate[J]. Plant Physiology, 1995, 109(2): 567-572.
[83] Ellis C, Turner J G. A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways inArabidopsisthalianaseeds and young seedlings[J]. Planta, 2002, 215(4): 549-556.
[84] Chen Q F, Dai L Y, Xiao S, et al. TheCOI1 andDFRgenes are essential for regulation of jasmonate-induced anthocyanin accumulation inArabidopsis[J]. Journal of Integrative Plant Biology, 2007, 49(9): 1370-1377.
[85] Devoto A, Ellis C, Magusin A, et al. Expression profiling revealsCOI1 to be a key regulator of genes involved in wound-and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions[J]. Plant Mol Biol, 2005, 58(4): 497-513.
[86] Ban Y, Honda C, Hatsuyama Y, et al. Isolation and functional analysis of aMYBtranscription factor gene that is a key regulator for the development of red coloration in apple skin[J]. Plant and cell physiology, 2007, 48(7): 958-970.
[87] Xie D X, Feys B F, James S, et al.COI1: anArabidopsisgene required for jasmonate-regulated defense and fertility[J]. Science, 1998, 280(5366): 1091-1114.
[88] Xu L, Liu F, Lechner E, et al. TheSCFCOI1 ubiquitin-ligase complexes are required for jasmonate response inArabidopsis[J]. Plant Cell, 2002, 14(8): 1919-1935.
[89] Qi T, Song S, Ren Q, et al. The Jasmonate-ZIM-domain proteins interact with theWD-Repeat/bHLH/MYBcomplexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation inArabidopsisthaliana[J]. Plant Cell, 2011, 23(5): 1795-1814.
(责任编辑:曾小军)
Summary of Regulation of Anthocyanin Synthesis by Light, Sugar and Hormone
WANG Hai-zhu1,2, XU Qi-jiang1,2, YAN Hai-fang2*
(1. State Key Laboratory of Tree Genetics and Breeding, Northeast Forestry University, Harbin 150040, China;2. College of Life Sciences, Northeast Forestry University, Harbin 150040, China)
Anthocyanin is a kind of ubiquitous flavonoids, and it exists in the vacuoles of epidermal cells of leaves, flowers, fruits and seeds of plants. Anthocyanin synthesis is not only regulated by structural genes and transcription factors, but also influenced by light, temperature, sugar, hormone, pH-value, and other factors. In this paper, the research progresses in the effects of light, sugar and hormone on anthocyanin biosynthesis pathway were reviewed.
Sugar; Light; Hormone; Anthocyanin synthesis
2016-03-11
中央高校基本科研业务费专项资金(DL12CA10、2572014EA03-01);中央高校基本科研业务费专项资金项目(2572014EA03);
王海竹(1991─),女,辽宁大连人,硕士研究生,研究方向:植物发育生物学和分子生物学。*通讯作者:闫海芳。
Q946.8
A
1001-8581(2016)09-0035-07
林木遗传育种国家重点实验室创新项目(2013A06、2013B010);黑龙江省博士后科研启动金(LBH-Q14011)。
