Advances and perspectives in the application of CRISPR/Cas9 in insects
2016-09-14LeiCHENGuiWANGYaNanZHUHuiXIANGWenWANGStateKeyLaboratoryofGeneticResourcesEvolutionKunmingInstituteofZoologyChineseAcademyofSciencesKunmingYunnan650ChinaUniversityofChineseAcademyofSciencesBeijing0009ChinaCollegeofHet
Lei CHEN, Gui WANG, Ya-Nan ZHU, Hui XIANG, Wen WANG,*State Key Laboratory of Genetic Resources & Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming Yunnan 650, ChinaUniversity of Chinese Academy of Sciences, Beijing 0009, ChinaCollege of Hetao, Bayannaoer Inner Mongolia 05000, ChinaFaculty of Life Science and Technology, Kunming University of Science and Technology, Kunming Yunnan 650, China
Advances and perspectives in the application of CRISPR/Cas9 in insects
Lei CHEN1,2,#, Gui WANG3,#, Ya-Nan ZHU1,4, Hui XIANG1, Wen WANG1,*
1State Key Laboratory of Genetic Resources & Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming Yunnan 650223, China
2University of Chinese Academy of Sciences, Beijing 100049, China
3College of Hetao, Bayannaoer Inner Mongolia 015000, China
4Faculty of Life Science and Technology, Kunming University of Science and Technology, Kunming Yunnan 650223, China
ABSTRACT
Insects compose more than half of all living organisms on earth, playing essential roles in global ecosystems and forming complex relationships with humans. Insect research has significant biological and practical importance. However, the application of genetic manipulation technology has long been restricted to several model insects only, such as gene knockout in Drosophila, which has severely restrained the development of insect biology research. Recently, with the increase in the release of insect genome data and the introduction of the CRISPR/Cas9 system for efficient genetic modification, it has been possible to conduct meaningful functional studies in a broad array of insect species. Here, we summarize the advances in CRISPR/Cas9 in different insect species, discuss methods for its promotion, and consider its application in future insect studies. This review provides detailed information about the application of the CRISPR/Cas9 system in insect research and presents possible ways to improve its use in functional studies and insect pest control.
CRISPR/Cas9; Gene editing; Insect transgenesis
INTRODUCTION
Insects compose more than half of all living organisms on earth and play essential roles in the world's ecosystems. They serve as pollinators, herbivores, and predators, and have the capacity to alter the rate and direction of energy and matter fluxes in ways that potentially affect global processes. Insects also exhibit complex relationships with humans. Agricultural pests, such as the diamondback moth and cotton bollworm, feed on plants and damage crops, with an estimated 10%-16% of global crop production lost to insect pests (Chakraborty & Newton, 2011). Furthermore,insects can be carriers of human disease. For example, Anopheles mosquitoes are vectors of malaria, an infectious and deadly disease threatening nearly half of the world's population according to the World Health Organization (World Health Organization, 2014). Conversely, insects such as silkworm and honey bees are economically and agriculturally important to human society, while social insects are ideal models for researching social behavior. Considering the important roles of insects in ecology and human life, insect research has great biological and practical significance.1
Nowadays, high-throughput genome sequencing technology has been used to decipher the genomes of many insects. Published genomes have gone far beyond model organisms such as Drosophila, now covering moths, butterflies, beetles and Hymenoptera social insects. These data not only provide the raw materials for further comparative and evolutionary genomic studies, but also bring about efficient gene function identification through newly emerged genome editing technologies, resulting in breakthroughs in the genetic manipulation of non-model organisms.
Traditional gene knock-in in insects was initially limited to transposon-based transgenic technologies, while the application of gene knockout was restricted to Drosophila species. Nowadays,however, newly emerged genome editing approaches, i.e., ZFN,TALEN and especially CRISPR/Cas9, have allowed for the efficient extension of genetic modification of non-model organisms and functional testing of candidate domesticatedgenes, as well as candidate genes in life-environment interactions (Chen et al., 2014). Accompanied by high-throughput sequencing platforms, CRISPR/Cas9 has promising impact on evolutionary and ecological research, as well as on elucidating the gap between DNA sequences and phenotypes. Here, we briefly introduce the development of insect genetic manipulation technology, with a focus on the CRISPR/Cas9 system in insects. We first summarize the advances of CRISPR/Cas9 in different insect species, and then discuss methods for its promotion and prospective application in future insect studies.
DEVELOPMENT OF GENETIC MANIPULATION IN INSECTS
Genetic manipulation technologies in insects have been developed over the last 30 years. Combined with recent progress, these technologies can be classified into three stages of development based on their function mechanism, efficiency,and accuracy. The first stage was transposon-based transgenesis. The first successful attempt to transfer exogenous genes into insects was conducted in Drosophila melanogaster by P-element transposon (Rubin & Spradling,1982). Unfortunately, the P-element was not applicable in nondrosophilid insects because of host-specific co-factor requirements (Rio & Rubin, 1988). Since then, however, four other transposons (mariner, Minos, Hermes and piggyBac)have been explored for the genetic transformation of nondrosophilid insects. Among them, universal vector piggyBac has been widely and successfully applied in the transgenesis of several insect species such as Bombyx mori (Tamura et al.,2000), butterfly (Marcus et al., 2004), and diamondback moth (Martins et al., 2012). Based on such studies, upgraded transgenesis technologies were developed, combined with the GAL4/UAS system as well as site-specific recombinases and integrases like Cre, FLP, and ΦC31 (Venken & Bellen, 2007). Nevertheless, transposons have remained limited in their utility,primarily due to integration randomness, low transformation frequency, integrated sequence instability, and limited carrying capacity. The second development stage of gene editing technologies such as ZFN (Zinc Finger Nuclease) and TALEN (Transcription Activator-Like Effector Nuclease), which are both composed of a specific DNA recognition protein (ZF or TALE)and DNA excision protein (FokI) (Bibikova et al., 2003;Bogdanove & Voytas, 2011) expanded genetic modifications beyond model organisms. Compared with transposon-based technologies, ZFN and TALEN improved the efficiency and precision of targeted genetic manipulations, and have been successfully applied in Drosophila (Gratz et al., 2013), B. mori (Wang et al., 2013), and mosquitoes (Dong et al., 2015). However, certain intrinsic drawbacks have restricted their wide utility. ZFN has limited target sites due to the three-nucleotide recognition model, and is expensive and difficult to assemble,whereas TALEN involves cumbersome procedures and a large protein that can be difficult to efficiently deliver into all cells. The third and most promising stage of advancement has been the CRISPR/Cas9 system (Clustered Regularly Interspaced Short Palindromic Repeats). The CRISPR/Cas9 system acts via a ribonucleoprotein complex, where the target recognition lobe of Cas9 directs specific binding to target DNA through interacting with homologous sgRNA and the excision lobe cuts the DNA (Nishimasu et al., 2014). It is considered a revolutionary technology with high efficiency and accuracy and applicable for a wide range of species. Many studies have reported on the application, evaluation, and improvement of the CRISPR/Cas9 system (Gaj et al., 2013; Hsu et al., 2014; Shalem et al., 2015). Here, we emphasize the application and potential of the CRISPR/Cas9 system in insects.
CURRENT APPLICATION OF THE CRISPR/CAS9 SYSTEM IN INSECTS
To date, the CRISPR/Cas9 system has been successfully applied in several insect species, including Drosophila, silkworm,mosquito, and butterfly (Table 1). Here, we summarize the advances of this technology in these insects, respectively.
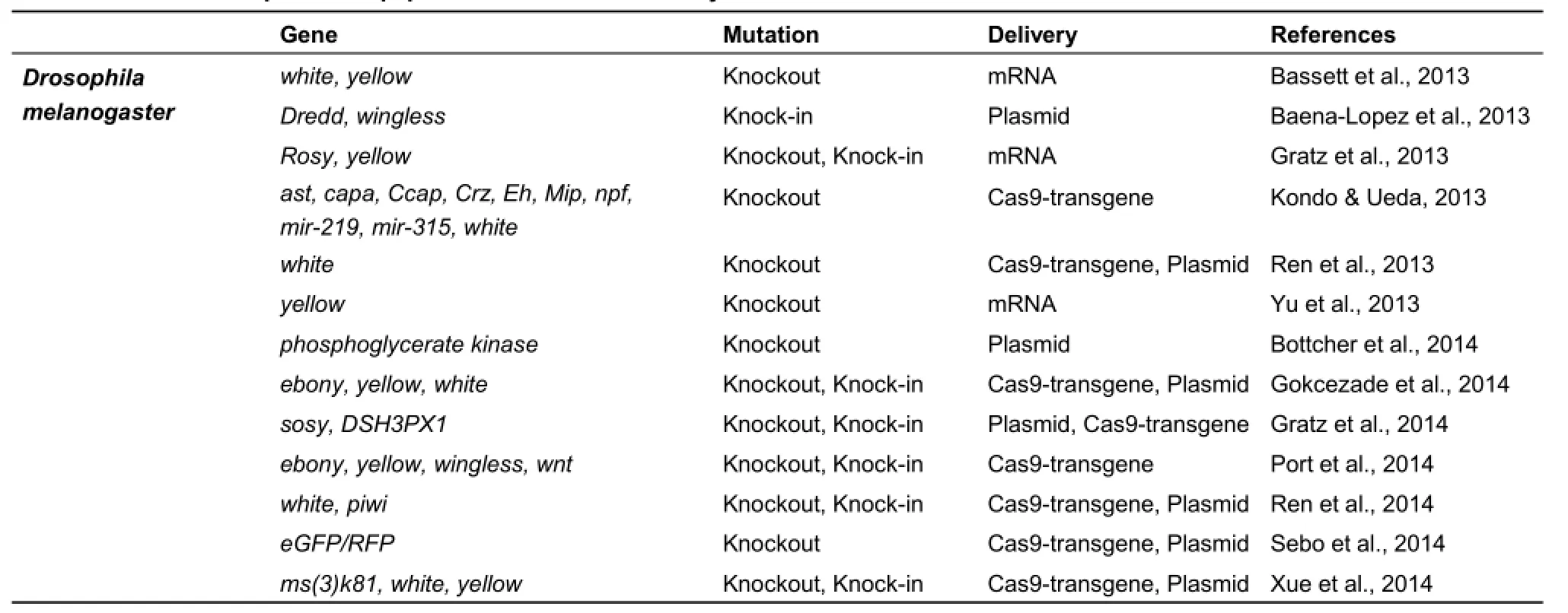
Table 1 Collections of published papers on the CRISPR/Cas9 system in insects
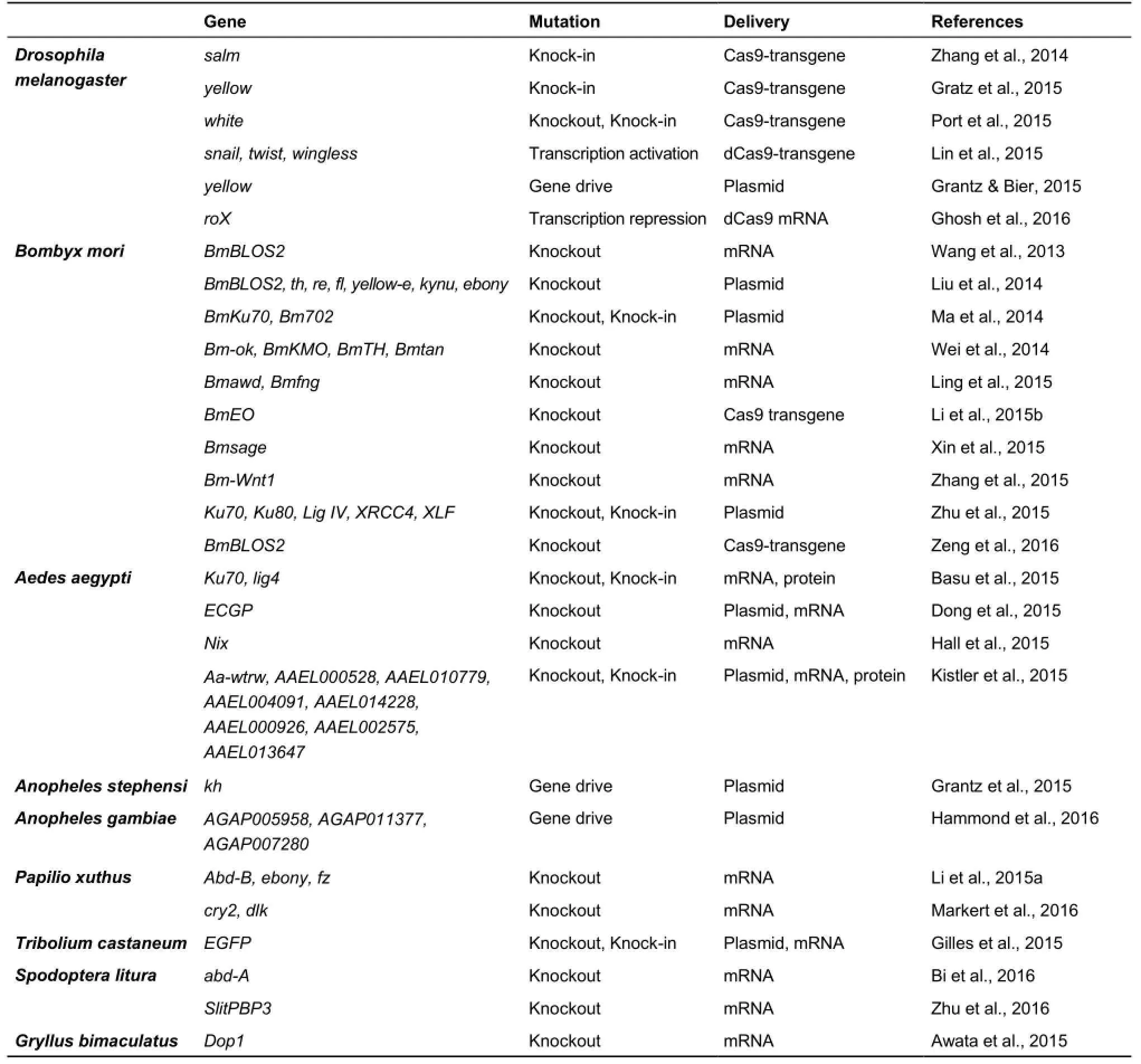
Continued
Drosophila
The first reported knockout application of CRISPR/Cas9 in insects was carried out in Drosophila melanogaster, in which mutations were introduced to the yellow gene (Gratz et al.,2013). Subsequently, significant research has been conducted on the development of the CRISPR/Cas9 system in Drosophila. Generally, there are four strategies to establish CRISPR/Cas9 system in Drosophila, depending on the Cas9 and sgRNA delivery methods. Firstly, Cas9 and sgRNA are both constructed into plasmids. In a pilot study, Gratz et al. (2013)constructed two plasmids expressing the Cas9 protein driven by the Hsp70 promoter and sgRNA driven by a U6 promoter, respectively. However, the efficiency of mutagenesis was relatively low (5.9%) due to inefficient error-prone nonhomologous end-joining (NHEJ) effects. Additionally, Cas9 and sgRNA could be merged into one single vector, which resulted in a mutagenesis rate>10% (Gokcezade et al., 2014). The second strategy was adopted by two research groups independently in which transcribed Cas9 mRNA and sgRNA were co-injected into early stage embryos in vitro (Bassett et al., 2013; Yu et al.,2013). More than 80% of injected flies presented mosaic expression of the yellow gene and another six genes showed 36%-80% mutagenesis, implying high efficiency. However, the high mutation rate relied on high concentrations of Cas9/sgRNA,resulting in an adult survival rate of ≤3% (Bassett et al., 2013).Conversely, reducing the concentration of Cas9/sgRNA mRNA resulted in the dramatic reduction in the proportion of mosaic adults from 86% to 10% (Bassett et al., 2013). The third strategy was established by crossing two stable transgenic fly strains: one specifically expressing the Cas9 protein in germline cells, the other ubiquitously expressing sgRNA. Expression of Cas9 driven by the nanos promoter and expression of sgRNA under the U6 promoter obtained highly efficient (average of 57%, reaching 90%) mutagenesis by crossing these two transgenic strains (Kondo & Ueda, 2013). This robust strategy generated much higher mutagenesis rate than the previous transient expression of Cas9 and sgRNAs, but was limited by the time spent establishing sgRNA transgenic fly stocks. The last strategy involved the injection of sgRNA expressing vectors into early stage embryos of germline-expressed Cas9 transgenic strains. This strategy reduced the complexity of the two-component injection scheme to one component. Previous studies constructed Cas9 transgenic strains by expressing the Cas9 protein driven by vasa (Sebo et al., 2014) or nanos (Ren et al., 2013) promoters. Sebo et al. (2014) observed a high ratio (up to 71%) of injected flies harboring gene mutations, although a significant portion (up to 68%) was infertile. Ren et al. (2013)achieved a higher rate of mutagenesis (93.3%) and fertility (81%) in G0 flies. As to promoter selection, different U6 promoters have been used to drive sgRNA, with U6: 3 demonstrated to have the strongest ability of generating mutagenesis in both somatic cells and germlines (Port et al.,2014). A new CR7T promoter was developed to efficiently drive sgRNA, and gave rise to high mutagenesis rates (Xue et al.,2014).
In addition to knockout technology, efficient and specific CRISPR/Cas9-mediated knock-in procedures have greatly expanded the application of CRISPR by facilitating complex genome modifications. Precise incorporation of exogenous DNA can be achieved through homology-directed repair (HR)by providing donor DNA templates. Previous studies have successfully injected donor DNA into embryos transgenic containing both Cas9 and sgRNA cascade (Port et al., 2014),donor DNA with sgRNA-encoding plasmids into transgenic Cas9 embryos (Gratz et al., 2014; Ren et al., 2013; Xue et al.,2014; Zhang et al., 2014), and donor plasmids together with a plasmid-encoding Cas9 and sgRNA into non-transgenic embryos (Gokcezade et al., 2014).
Silkworm
The first successful application of the CRISPR/Cas9 system was reported for B. mori using the BmBLOS2 gene as a target (Wang et al., 2013). Two 23 bp gRNAs were designed to direct the Cas9 endonuclease specifically to two sites separated by 3.48 kb of BmBLOS2, with a high mutagenesis rate obtained for each site (94% and 95.6%) and large deletions achieved by injection of in vitro transcribed Cas9 mRNA and sgRNAs (Wang et al., 2013). Similarly, four more genes (Bm-ok, BmKMO,BmTH and Bmtan) were altered at specific sites by the direct microinjection of specific sgRNA and Cas9 mRNA into embryos,with mutation frequencies of 16.7%-35.0% observed in the injected generation (Wei et al., 2014). Ma et al. (2014) established a system of two expression vectors for Cas9 and sgRNA separately, generating the heritable site-directed edition of Bmku70 in B. mori. This same system was also used to induce multiplex genome editing of six genes in BmNs cell lines simultaneously (Liu et al., 2014). In addition to knockout, HR introduced knock-in has also been reported. Ma et al. (2014)found that HR frequency increased in Bmku70 knocked-out B. mori. Additionally, Zhu et al. (2015) knocked out the factors Ku70, Ku80, Lig IV, XRCC4 and XLF in NHEJ using the CRISPR/Cas9 system, which increased the activities of HR up to 7-fold in silkworm cells. Further efforts have also been made to improve the CRISPR/Cas9 system in recent years. A U6 promoter from B. mori was shown to effectively drive sgRNA initiated with nucleotide bases (N20NGG) and induce mutations in vitro and in vivo (Zeng et al., 2016).
The highly efficient CRISPR/Cas9 system has also been applied in gene functional analysis of the miR-2 cluster by knockout of miR-2 targeted Bmawd and Bmfng genes in silkworm (Ling et al., 2015). Furthermore, functional analysis of the BmWnt1 gene using the CRISPR/Cas9 system generated a large deletion of 18 kb, resulting in severe developmental defects during embryogenesis, comparable to previously reported RNAi studies (Zhang et al., 2015). The transgenic CRISPR/Cas9 system also somatically mutated BmEO and extended the duration of the final instar larval stage of silkworm (Li et al., 2015b). Combined with Gal4/UAS overexpression and RNA-seq analysis, CRISPR/Cas9 provided insights into BmEO ecdysone regulation of tissue degeneration during metamorphosis (Li et al., 2015b). Germline mutation of Bmsage was introduced using the Cas9/sgRNA system, leading to poorly developed silk glands and absent middle and posterior silk glands (MSG and PSG) (Xin et al., 2015), implying that the Bmsage gene participates in the development of these glands at the embryonic stage.
Mosquito
Aedes aegypti is the principal vector for important arboviruses such as yellow fever, dengue, and chikungunya viruses, which cause significant impacts on human health. The CRISPR/Cas9 system has been used in in vivo gene disruptions of ECFP in ECFP transgenic A. aegypti lines (Dong et al., 2015). Injection of in vitro transcribed Cas9 mRNA and sgRNA into embryos introduced mutations with 5.5% knockout efficiency (Dong et al.,2015). Kistler et al. (2015) increased the CRISPR/Cas9 mutation rate up to 24% by identifying active sgRNAs and proper Cas9 mRNA concentrations in five A. aegypti genes. Basu et al. (2015) promoted the effectiveness of editing in A. aegypti up to 90%, in which a large cohort of sgRNAs were evaluated in early embryos, with only highly ranked candidates going forward to germline-based experiments. It should be noted that injection of plasmids encoding Cas9 and sgRNA did not increase observed mutations, which might result from the weak promoters used to drive Cas9 and sgRNAs or intrinsically low efficiency of the CRISP/Cas9 system in A. aegypti (Dong et al., 2015; Kistler et al., 2015). HR-based integration of a transgene in A. aegypti was also obtained using the CRISPR/Cas9 system, with a transformation rate of 2.1%obtained (Basu et al., 2015). The CRISPR/Cas9 system has also been applied in the functional study of Nix, the first insect M factor. Knockout of Nix resulted in largely feminized genetic males, providing an opportunity to characterize the remaining genes and interactions in the A. aegypti sex-determination pathway (Hall et al., 2015).
Butterfly
Recent application of CRISPR/Cas9 was found to induce somatic mutations of the Abd-B gene with high efficiency (92.5%) in swallowtail butterfly, Papilio xuthus (Li et al., 2015a). Furthermore, a highly efficient, heritable gene knockout at two clock gene loci, cry2 and clk, was reported in the monarch butterfly, Danaus plexippus (Markert et al., 2016), in which 50% of larvae presented indels at rates ranging from 3%-28%. The knockout for clk helped define its critical function in encoding a transcriptional activator of the circadian clock used by monarchs during migration.
Other insects
The CRISPR/Cas9 system has shown great potential in the genetic editing of non-model species. Gilles et al. (2015)demonstrated that CRISPR technology effectively generated targeted knockouts, knock-ins, and deletions in Tribolium castaneum. Furthermore, CRISPR/Cas9 also induced efficient gene mutagenesis of the typically deficient phenotype of abd-A in Spodoptera litura, a serious agricultural pest worldwide, by direct injection of Cas9 mRNA and sgRNA into S. litura embryos (Bi et al., 2016). CRISPR/Cas9 has also been used in functional characterization of the SlitPBP3 gene in S. litura (Zhu et al., 2016). Furthermore, the Dop1 gene was knocked out in non-model Gryllus bimaculatus using the CRISPR/Cas9 system,resulting in mutants that were defective in aversive learning with sodium chloride punishment, but not appetitive learning with water or sucrose reward (Awata et al., 2015).
IMPROVEMENT OF THE CRISPR/CAS9 SYSTEMS IN INSECTS
Efficiency
Cas9 can efficiently generate knockout in insects via NHEJ, but the efficiency of precise sequence replacement via HR is substantially lower. Ma et al. (2014) attempted to promote knock-in efficiency by targeting BmKu70, which is involved in the NHEJ pathway. Several methods have been developed to increase knock-in efficiency in vivo, which might provide new methods for application in insects. Precise knock-in is usually accomplished by stimulating HR in the presence of a singlestranded oligodeoxynucleotide (ssODN) template, with ssODN-mediated knock-in achieved in Drosophila (Gratz et al., 2013;Liu et al., 2014), silkworm (Ma et al., 2014; Zhu et al., 2015),and mosquito (Kistler et al., 2015). Recently, the rational design of ssDNA donors to match first released DNA strands by the Cas9 protein increased the rate of HR by 60% (Richardson et al., 2016). In addition, the two-hit two-oligo (2H2OP) method composed of Cas9 and two sgRNAs generated knock-in of 200 kb sequences into a genome through HR (Yoshimi et al., 2016). However, HR is not the only pathway leading to precise knockins. The CRISPR/Cas9 system can also mediate knock-in of exogenous DNA based on the NHEJ repair pathway. In zebrafish, for example, the mutation rate was promoted up to 66%, much higher than the 1.5% mutation rate by HR (Auer et al., 2014a, b). Microhomology-mediated end-joining (MMEJ),another non-homologous DSB repair mechanism, uses microhomologous sequences (5-25 bp) for error-prone end-joining. Based on the CRISPR/Cas9 system, Nakade et al. (2014)developed precise integration into the target chromosome (CRIS-PITCh) system, which enabled the efficient integration of exogenous DNA in vivo. TALEN-mediated PITCh has also been shown to be quite effective in silkworm (Nakade et al., 2014).
Accuracy
The Cas9/sgRNA system has been successfully applied in both model and non-model insects to induce indel mutations through NHEJ or sequence-specific mutations through HR. However,potential off-target effects might result in unexpected indel mutations, especially when relying on NHEJ, thus increasing the complexity of analyzing mutants of interest (Cho et al., 2014;Fu et al., 2013; Hsu et al., 2013). Several methods can reduce off-target effects and promote targeting specificities. Firstly,introduction of a D10A or H840A mutation into the RuvC1- or HNH-like nuclease domains in Cas9 can generate a variant protein with single-stranded DNA cleavage (nickase) activity (Gasiunas et al., 2012; Jinek et al., 2012). Cas9 nickase (dCas9)with paired gRNAs properly positioned on the target DNA exhibits low off-target mutagenesis compared with wild-type Cas9 (Cho et al., 2014; Mali et al., 2013; Ran et al., 2013). In Drosophila, the application of Cas9 nickase and sgRNA pairs almost eliminated off-target effects when generating indel mutants (Ren et al., 2014). However, Cas9D10A was not as effective as Cas9 in replacing the entire coding sequence of piwi with two sgRNAs (Ren et al., 2014). Previous studies have also used sgRNA-guided dCas9 fused to FokI nuclease where two fused dCas9-FokI bind target sites at a defined distance apart, inducing DNA double-strand break after dimerization of the two monomers (Guilinger et al., 2014; Tsai et al., 2014). Furthermore, truncated sgRNAs (17-18 nt long) were found to decrease undesired mutagenesis at some off-target sites by 5,000-fold or more while maintaining on-target activities (Fu et al.,2014). However, this method is not robust and might not work on some targets, thus reducing the number of target sites in the genome. The architecture of the Cas9 protein can also be modified to reduce off-target effects. Kleinstiver et al. (2015)screened a D1135E mutation in the PAM-interacting domains by bacterial selection, which improved the specificity for offtarget sites without decreasing on-target activity. In addition,some studies decreased the energetics of interaction between the Cas9-sgRNA complex and target DNA, so that CRISPR/Cas9 might retain robust on-target activity, but have a diminished ability to cleave mismatched off-target sites. Three amino acids of SpCas9 (K848A/K1003A/R1060A) were engineered, which decreased off-target indel formation by at least 10-fold, without scarifying on-target activity (Slaymaker et al., 2016). In another study, four SpCas9 residues (N497A,R661A, Q695A, Q926A) were reformed to generate a SpCas9-HF1 protein, which reduced all or nearly all genome-wide offtarget effects to undetectable levels (Kleinstiver et al., 2016).
PROSPECTIVE APPLICATION IN INSECTS
The application of CRISPR/Cas9 in insects is still in the early stages. It has been intensely reformed for different applications in model animals, which might clarify prospective applications in insects.
Gene drive
Gene drive refers to the increase in the frequency of particular genes by bias inheritance. Based on the CRISPR/Cas9 genome editing system, a mutagenic chain reaction (MCR)method was developed to conduct gene drive in Drosophila (Gantz & Bier, 2015), in which MCR converted heterozygous mutations to homozygosity at the yellow locus in germlines with 96% homing efficiency by copying themselves onto the homologous chromosome through HR. This study demonstrated that MCR technology was highly efficient in Drosophila and could be applied in other insect species. Recently, MCR technology was also established in Anopheles stephensi, where eye color gene kh was targeted with 99.5% efficiency (Gantz et al., 2015). Similarly, in Anopheles gambiae,three female-sterility genes (AGAP005958, AGAP011377 and AGAP007280) were identified and triggered expected phenotypes with 91.4% to 99.6% efficiencies (Hammond et al.,2016). The reduction in female fertility has the potential to substantially reduce mosquito populations. These pilot experiments verify that the CRISPR-Cas9 gene drive system is a robust and valuable gene editing tool for functional genetic research in insects and in disease control by suppressing mosquito populations.
Regulation
The CRISPR/Cas9 system has the potential to activate or repress gene expression in vivo. CRISPR inference (CRISPRi)is a modified CRISPR/Cas9 system in which dCas9 paired with sgRNA can sterically hinder transcription at the sgRNA basepairing genomic locus (Qi et al., 2013). The CRISPRi framework has been used for systematic phenotypic analysis of essential genes in bacteria (Peters et al., 2016). Additionally,CRISPRi regulation can be used to achieve activation or repression by fusing dCas9 to activator or repressor modules (Gilbert et al., 2013; Gilles et al., 2015; Mali et al., 2013). However, modest levels of gene activation have limited potential application. An improved transcriptional regulator was obtained through the rational design of a tripartite activator,VP64-p65-rta (VPr) (Chavez et al., 2015). In another study,different Cas9 activator systems and their ability to induce robust gene expression were examined in human, mouse, and fly cell lines, with VPR, SAM, and Suntag activators found to be superior to VP64 (Chavez et al., 2016). Nevertheless, the direct fusing of dCas9 with effector proteins is constrained to only one direction of regulation. Zalatan et al. (2015) converted sgRNA to scaffold RNA (scRNA) by extending the sgRNA sequence with modular RNA domains (MS2, PP7 and com), thus enabling flexible and parallel programmable locus-specific regulation. Multiple genes can be regulated in different directions (activation or repression) simultaneously. Using CRISPRi, the coexpression of dCas9 and guide RNAs targeted the endogenous roX locus (long non-coding RNAs) in Drosophila cells, resulting in robust and specific knockdown of roX1 and roX2 RNAs (Ghosh et al., 2016).
DNA or RNA tracking
Live imaging systems for visualization of genome loci are essential for studying chromatin dynamics and nuclear localization, which is important for understanding cellar processes. The CRISPR/Cas9 system can be reformed as a live imaging system by tagging dCas9 proteins with enhanced green fluorescent protein (EGFP) (Chen et al., 2013). An essential feature of any live imaging system is the ability to visualize more than one locus at a time. By using dCas9 from three bacterial orthologues fused with different fluorescent proteins, dCas9 and sgRNAs were found to efficiently label several target loci in live human cells (Chen et al., 2016; Ma et al., 2015). Another more flexible method is extending sgRNAs with RNA domains (MS2 and PP7), which when co-expressed with a dCas9 can recruit fluorescently tagged RNA-binding proteins (MCP and PCP) to specific genomic sites (Fu et al.,2016). This allows for rapid, stable dual-color labelling. RNAs can also be tracked dynamically. Targeting RNAs with dCas9 is possible by providing PAM as part of an oligonucleotide (PAMmer) that hybridizes to the target RNA (O'Connell et al.,2014). Based on such a system, RNA-targeting Cas9 (RCas9)was generated with dCas9 fused with fluorescent protein (e.g. GFP), sgRNA and PAMmer, which recognized mRNAs in live cells while avoiding encoding DNA (Nelles et al., 2016).
CONCLUSIONS
The CRISPR/Cas9 system is a revolutionary tool for both prokaryotic and eukaryotic genetics. It has been preliminarily established and developed in model and non-model insects. Highly efficient knockout and knock-in experiments have been successfully conducted in model insects such as Drosophila and silkworm, and in non-model insects such as butterfly,mosquito, and beetle. Based on CRISPR/Cas9, several welldesigned systems have been developed, including gene drive and regulation and DNA/RNA tracking systems, which will have significant impact on functional studies and pest control. Researchers have improved the CRISPR/Cas9 system in insects, resulting in easier and more effective design and use. Successful modifications of CRISPR/Cas9 have been made in cells and model animals, implying that CRISPR/Cas9 has the potential for broad application and development in insects.
REFERENCES
Auer TO, Duroure K, Concordet JP, Del Bene F. 2014a. CRISPR/Cas9-mediated conversion of eGFP- into Gal4-transgenic lines in zebrafish. Nature Protocols, 9(12): 2823-2840.
Auer TO, Duroure K, De Cian A, Concordet JP, Del Bene F. 2014b. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homologyindependent DNA repair. Genome Research, 24(1): 142-153.
Awata H, Watanabe T, Hamanaka Y, Mito T, Noji S, Mizunami M. 2015.
Knockout crickets for the study of learning and memory: Dopamine receptor Dop1 mediates aversive but not appetitive reinforcement in crickets. Scientific Reports, 5: 15885.
Bassett AR, Tibbit C, Ponting CP, Liu JL. 2013. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Reports,4(1): 220-228.
Basu S, Aryan A, Overcash JM, Samuel GH, Anderson MaE, Dahlem TJ,Myles KM, Adelman ZN. 2015. Silencing of end-joining repair for efficient site-specific gene insertion after TALEN/CRISPR mutagenesis in Aedes aegypti. Proceedings of the National Academy of Sciences of the United States of America, 112(13): 4038-4043.
Bi HL, Xu J, Tan AJ, Huang YP. 2016. CRISPR/Cas9-mediated targeted gene mutagenesis in Spodoptera litura. Insect Science, 23(3): 469-477.
Bibikova M, Beumer K, Trautman JK, Carroll D. 2003. Enhancing gene targeting with designed zinc finger nucleases. Science, 300(5620): 764.
Bogdanove AJ, Voytas DF. 2011. TAL effectors: customizable proteins for DNA targeting. Science, 333(6051): 1843-1846.
Chakraborty S, Newton AC. 2011. Climate change, plant diseases and food security: an overview. Plant Pathology, 60(1): 2-14.
Chavez A, Tuttle M, Pruitt BW, Ewen-Campen B, Chari R, Ter-Ovanesyan D, Haque SJ, Cecchi RJ, Kowal EJK, Buchthal J, Housden BE, Perrimon N,Collins JJ, Church G. 2016. Comparison of Cas9 activators in multiple species. Nature Methods, 13(7): 563-567.
Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, P R Iyer E, Lin S, Kiani S, Guzman CD, Wiegand DJ, Ter-Ovanesyan D, Braff JL, Davidsohn N,
Housden BE, Perrimon N, Weiss R, Aach J, Collins JJ, Church GM. 2015. Highly efficient Cas9-mediated transcriptional programming. Nature Methods, 12(4): 326-328.
Chen BH, Hu J, Almeida R, Liu H, Balakrishnan S, Covill-Cooke C, Lim WA,Huang B. 2016. Expanding the CRISPR imaging toolset with Staphylococcus aureus Cas9 for simultaneous imaging of multiple genomic loci. Nucleic Acids Research, 44(8): e75.
Chen BH, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J,Blackburn EH, Weissman JS, Qi LS, Huang B. 2013. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell, 155(7): 1479-1491.
Chen L, Tang LY, Xiang H, Jin LJ, Li QY, Dong Y, Wang W, Zhang GJ. 2014. Advances in genome editing technology and its promising application in evolutionary and ecological studies. GigaScience, 3: 24.
Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. 2014. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Research, 24(1): 132-141.
Dong SZ, Lin JY, Held NL, Clem RJ, Passarelli AL, Franz AWE. 2015. Heritable CRISPR/Cas9-mediated genome editing in the yellow fever mosquito, Aedes aegypti. PLoS One, 10(3): e0122353.
Fu Y, Rocha PP, Luo VM, Raviram R, Deng Y, Mazzoni EO, Skok JA. 2016. CRISPR-dCas9 and sgRNA scaffolds enable dual-colour live imaging of satellite sequences and repeat-enriched individual loci. Nature Communications, 7: 11707.
Fu YF, Sander JD, Reyon D, Cascio VM, Joung JK. 2014. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nature Biotechnology, 32(3): 279-284.
Fu YF, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. 2013. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature Biotechnology, 31(9): 822-826.
Gaj T, Gersbach CA, Barbas CF. 2013. ZFN, TALEN, and CRISPR/Casbased methods for genome engineering. Trends in Biotechnology, 31(7):397-405.
Gantz VM, Bier E. 2015. The mutagenic chain reaction: A method for converting heterozygous to homozygous mutations. Science, 348(6233):442-444.
Gantz VM, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, Bier E,James AA. 2015. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proceedings of the National Academy of Sciences of the United States of America, 112(49): E6736-E6743.
Gasiunas G, Barrangou R, Horvath P, Siksnys V. 2012. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences of the United States of America, 109(39): E2579-E2586.
Ghosh S, Tibbit C, Liu JL. 2016. Effective knockdown of Drosophila long non-coding RNAs by CRISPR interference. Nucleic Acids Research, 44(9):e84.
Gilbert LA, Larson MH, Morsut L, Liu ZR, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA. 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell, 154(2): 442-451.
Gilles AF, Schinko JB, Averof M. 2015. Efficient CRISPR-mediated gene targeting and transgene replacement in the beetle Tribolium castaneum. Development, 142(16): 2832-2839.
Gokcezade J, Sienski G, Duchek P. 2014. Efficient CRISPR/Cas9 plasmids for rapid and versatile genome editing in Drosophila. G3: Genes Genomes Genetics, 4(11): 2279-2282.
Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM,O'connor-Giles KM. 2014. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics, 196(4): 961-971.
Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O'connor-Giles KM. 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics, 194(4):1029-1035.
Guilinger JP, Thompson DB, Liu DR. 2014. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nature Biotechnology, 32(6): 577-582.
Hall AB, Basu S, Jiang XF, Qi YM, Timoshevskiy VA, Biedler JK,Sharakhova MV, Elahi R, Anderson MaE, Chen XG, Sharakhov IV, Adelman ZN, Tu ZJ. 2015. A male-determining factor in the mosquito Aedes aegypti. Science, 348(6240): 1268-1270.
Hammond A, Galizi R, Kyrou K, Simoni A, Siniscalchi C, Katsanos D,Gribble M, Baker D, Marois E, Russell S, Burt A, Windbichler N, Crisanti A,Nolan T. 2016. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nature Biotechnology, 34(1): 78-83.
Hsu PD, Lander ES, Zhang F. 2014. Development and applications ofCRISPR-Cas9 for genome engineering. Cell, 157(6): 1262-1278.
Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li YQ,Fine EJ, Wu XB, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature Biotechnology, 31(9): 827-832.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science, 337(6096): 816-821.
Kistler KE, Vosshall LB, Matthews BJ. 2015. Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. Cell Reports, 11(1): 51-60.
Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng ZL,Joung JK. 2016. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature, 529(7587): 490-495.
Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng ZL,Gonzales APW, Li ZY, Peterson RT, Yeh JRJ, Aryee MJ, Joung JK. 2015. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature,523(7561): 481-485.
Kondo S, Ueda R. 2013. Highly improved gene targeting by germlinespecific Cas9 expression in Drosophila. Genetics, 195(3): 715-721.
Li XY, Fan DD, Zhang W, Liu GC, Zhang L, Zhao L, Fang XD, Chen L,Dong Y, Chen Y, Ding Y, Zhao RP, Feng MJ, Zhu YB, Feng Y, Jiang XT, Zhu DY, Xiang H, Feng XK, Li SC, Wang J, Zhang GJ, Kronforst MR, Wang W. 2015a. Outbred genome sequencing and CRISPR/Cas9 gene editing in butterflies. Nature Communications, 6: 8212.
Li ZQ, You L, Zeng BS, Ling L, Xu J, Chen X, Zhang ZJ, Palli SR, Huang YP, Tan AJ. 2015b. Ectopic expression of ecdysone oxidase impairs tissue degeneration in Bombyx mori. Proceedings of the Royal Society of London B: Biological Sciences, 282(1809): 20150513.
Ling L, Ge X, Li ZQ, Zeng BS, Xu J, Chen X, Shang P, James AA, Huang YP, Tan AJ. 2015. MiR-2 family targets awd and fng to regulate wing morphogenesis in Bombyx mori. RNA Biology, 12(7): 742-748.
Liu YY, Ma SY, Wang XG, Chang JS, Gao J, Shi R, Zhang JD, Lu W, Liu Y,Zhao P, Xia QY. 2014. Highly efficient multiplex targeted mutagenesis and genomic structure variation in Bombyx mori cells using CRISPR/Cas9. Insect Biochemistry and Molecular Biology, 49: 35-42.
Ma HH, Naseri A, Reyes-Gutierrez P, Wolfe SA, Zhang SJ, Pederson T. 2015. Multicolor CRISPR labeling of chromosomal loci in human cells. Proceedings of the National Academy of Sciences of the United States of America, 112(10): 3002-3007.
Ma SY, Chang JS, Wang XG, Liu YY, Zhang JD, Lu W, Gao J, Shi R, Zhao P, Xia QY. 2014. CRISPR/Cas9 mediated multiplex genome editing and heritable mutagenesis of BmKu70 in Bombyx mori. Scientific Reports, 4:4489.
Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L,Church GM. 2013. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nature Biotechnology, 31(9): 833-838.
Marcus JM, Ramos DM, Monteiro A. 2004. Germline transformation of the butterfly Bicyclus anynana. Proceedings of the Royal Society of London B: Biological Sciences, 271(Suppl 5): S263-S265.
Markert MJ, Zhang Y, Enuameh MS, Reppert SM, Wolfe SA, Merlin C. 2016. Genomic access to monarch migration using TALEN and CRISPR/Cas9-mediated targeted mutagenesis. G3: Genes Genomes Genetics, 6(4): 905-915.
Martins S, Naish N, Walker AS, Morrison NI, Scaife S, Fu G, Dafa'alla T,Alphey L. 2012. Germline transformation of the diamondback moth, Plutella xylostella L., using the piggyBac transposable element. Insect Molecular Biology, 21(4): 414-421.
Nakade S, Tsubota T, Sakane Y, Kume S, Sakamoto N, Obara M, Daimon T,Sezutsu H, Yamamoto T, Sakuma T, Suzuki KT. 2014. Microhomologymediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nature Communications, 5:5560.
Nelles DA, Fang MY, O'Connell MR, Xu JL, Markmiller SJ, Doudna JA, Yeo GW. 2016. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell, 165(2): 488-496.
Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N,Ishitani R, Zhang F, Nureki O. 2014. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell, 156(5): 935-949.
O'Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M,Doudna JA. 2014. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature, 516(7530): 263-266.
Peters JM, Colavin A, Shi HD, Czarny TL, Larson MH, Wong S, Hawkins JS, Lu CHS, Koo BM, Marta E, Shiver AL, Whitehead EH, Weissman JS,Brown ED, Qi LS, Huang KC, Gross CA. 2016. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell, 165(6): 1493-1506.
Port F, Chen HM, Lee T, Bullock SL. 2014. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proceedings of the National Academy of Sciences of the United States of America, 111(29): E2967-E2976.
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. 2013. Repurposing CRISPR as an RNA-guided platform for sequencespecific control of gene expression. Cell, 152(5): 1173-1183.
Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. 2013. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell, 154(6):1380-1389.
Ren XJ, Sun J, Housden BE, Hu YH, Roesel C, Lin SL, Liu LP, Yang ZH,Mao DC, Sun LZ, Wu QJ, Ji JY, Xi JZ, Mohr SE, Xu J, Perrimon N, Ni JQ. 2013. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proceedings of the National Academy of Sciences of the United States of America, 110(47): 19012-19017.
Ren XJ, Yang ZH, Mao DC, Chang Z, Qiao HH, Wang X, Sun J, Hu Q, Cui Y, Liu LP, Ji JY, Xu J, Ni JQ. 2014. Performance of the Cas9 nickase system in Drosophila melanogaster. G3: Genes Genomes Genetics, 4(10):1955-1962.
Richardson CD, Ray GJ, Dewitt MA, Curie GL, Corn JE. 2016. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nature Biotechnology, 34(3):339-344.
Rio DC, Rubin GM. 1988. Identification and purification of a Drosophila protein that binds to the terminal 31-base-pair inverted repeats of the P transposable element. Proceedings of the National Academy of Sciences of the United States of America, 85(23): 8929-8933.
Rubin GM, Spradling AC. 1982. Genetic transformation of Drosophila with transposable element vectors. Science, 218(4570): 348-353.
Sebo ZL, Lee HB, Peng Y, Guo Y. 2014. A simplified and efficient germline-
specific CRISPR/Cas9 system for Drosophila genomic engineering. Fly,8(1): 52-57.
Shalem O, Sanjana NE, Zhang F. 2015. High-throughput functional genomics using CRISPR-Cas9. Nature Reviews Genetics, 16(5): 299-311.
Slaymaker IM, Gao LY, Zetsche B, Scott DA, Yan WX, Zhang F. 2016. Rationally engineered Cas9 nucleases with improved specificity. Science,351(6268): 84-88.
Tamura T, Thibert C, Royer C, Kanda T, Abraham E, Kamba M, Komoto N,Thomas JL, Mauchamp B, Chavancy G, Shirk P, Fraser M, Prudhomme JC,Couble P. 2000. Germline transformation of the silkworm Bombyx mori L. using a piggyBac transposon-derived vector. Nature Biotechnology, 18(1):81-84.
Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. 2014. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nature Biotechnology, 32(6):569-576.
Venken KJT, Bellen HJ. 2007. Transgenesis upgrades for Drosophila melanogaster. Development, 134(20): 3571-3584.
Wang YQ, Li ZQ, Xu J, Zeng BS, Ling L, You L, Chen YZ, Huang YP, Tan AJ. 2013. The CRISPR/Cas system mediates efficient genome engineering in Bombyx mori. Cell Research, 23(12): 1414-1416.
Wei W, Xin HH, Roy B, Dai JB, Miao YG, Gao GJ. 2014. Heritable genome editing with CRISPR/Cas9 in the silkworm, Bombyx mori. PLoS One, 9(7):e101210.
World Health Organization. 2014. World Malaria Report 2014.
Xin HH, Zhang DP, Chen RT, Cai ZZ, Lu Y, Liang S, Miao YG. 2015. Transcription factor bmsage plays a crucial role in silk gland generation in silkworm, Bombyx mori. Archives of Insect Biochemistry and Physiology,90(2): 59-69.
Xue ZY, Ren MD, Wu MH, Dai JB, Rong YS, Gao GJ. 2014. Efficient gene knock-out and knock-in with transgenic Cas9 in Drosophila. G3: Genes Genomes Genetics, 4(5): 925-929.
Yoshimi K, Kunihiro Y, Kaneko T, Nagahora H, Voigt B, Mashimo T. 2016. ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nature Communications, 7: 10431.
Yu ZS, Ren MD, Wang ZX, Zhang B, Rong YS, Jiao RJ, Gao GJ. 2013. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics, 195(1): 289-291.
Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M,Tsai JC, Weissman JS, Dueber JE, Qi LS, Lim WA. 2015. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell, 160(1-2): 339-350.
Zeng BS, Zhan S, Wang YQ, Huang YP, Xu J, Liu Q, Li ZQ, Huang YP, Tan AJ. 2016. Expansion of CRISPR targeting sites in Bombyx mori. Insect Biochemistry and Molecular Biology, 72: 31-40.
Zhang X, Koolhaas WH, Schnorrer F. 2014. A versatile two-step CRISPR-and RMCE-based strategy for efficient genome engineering in Drosophila. G3: Genes Genomes Genetics, 4(12): 2409-2418.
Zhang ZJ, Aslam AFM, Liu XJ, Li MW, Huang YP, Tan AJ. 2015. Functional analysis of Bombyx Wnt1 during embryogenesis using the CRISPR/Cas9 system. Journal of Insect Physiology, 79: 73-79.
Zhu GH, Xu J, Cui Z, Dong XT, Ye ZF, Niu DJ, Huang YP, Dong SL. 2016. Functional characterization of SlitPBP3 in Spodoptera litura by CRISPR/Cas9 mediated genome editing. Insect Biochemistry and Molecular Biology, 75: 1-9.
Zhu L, Mon H, Xu J, Lee JM, Kusakabe T. 2015. CRISPR/Cas9-mediated knockout of factors in non-homologous end joining pathway enhances gene targeting in silkworm cells. Scientific Reports, 5: 18103.
10.13918/j.issn.2095-8137.2016.4.220
01 July 2016; Accepted: 13 July 2016
Foundation items: This work was supported by a 973 program (2013CB835200) to Wen WANG; a key grant of West Light Foundation of the Chinese Academy of Sciences to Hui XIANG
*Corresponding author, E-mail: wwang@mail.kiz.ac.cn
#Authors contributed equally to this work
杂志排行
Zoological Research的其它文章
- Early embryonic development and transplantation in tree shrews
- Molecular cloning and anti-HIV-1 activities of APOBEC3s from northern pig-tailed macaques (Macaca leonina)
- Research proceedings on amphibian model organisms
- Modeling postpartum depression in rats: theoretic and methodological issues
- Application of the genome editing tool CRISPR/Cas9 in non-human primates
- Generation of genetically modified mice using CRISPR/Cas9 and haploid embryonic stem cell systems
