The Australasian frog family Ceratobatrachidae in China, Myanmar and Thailand: discovery of a new Himalayan forest frog clade
2016-03-22FangYANKeJIANGKaiWANGJieQiongJINChatmongkonSUWANNAPOOM4ChengLIJensVINDUMRafeBROWNJingCHE
Fang YAN, Ke JIANG, Kai WANG, Jie-Qiong JIN, Chatmongkon SUWANNAPOOM4,, Cheng LI, Jens V. VINDUM, Rafe M. BROWN, Jing CHE,2,*
1State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming Yunnan 650223, China
The Australasian frog family Ceratobatrachidae in China, Myanmar and Thailand: discovery of a new Himalayan forest frog clade
Fang YAN1,2,#, Ke JIANG1,2,#, Kai WANG1,3, Jie-Qiong JIN1, Chatmongkon SUWANNAPOOM4,1, Cheng LI5, Jens V. VINDUM6, Rafe M. BROWN7, Jing CHE1,2,*
1State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming Yunnan 650223, China
2Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences, Yezin, Nay Pyi Taw 05282, Myanmar
3Sam Noble Oklahoma Museum of Natural History and Department of Biology, University of Oklahoma, Norman OK 73072-7029, U.S.A.4School of Agriculture and Natural Resources, University of Phayao, Phayao 56000, Thailand
5Imaging Biodiversity Expedition, Beijing 100107, China
6Department of Herpetology, California Academy of Sciences, California 94118, U.S.A.
7Biodiversity Institute and Department of Ecology and Evolutionary Biology, 1345 Jayhawk Blvd, University of Kansas, Lawrence KS 66045, U.S.A.
Received: 27 October 2015; Accepted: 15 December 2015
Foundation items: This study was supported by the Ministry of Science and Technology of China (2014FY210200, 2011FY120200), the program of Chinese Academy of Sciences (2015CASEABRI002), and the Animal Branch of the Germplasm Bank of Wild Species of Chinese Academy of Sciences (the Large Research Infrastructure Funding) to JC; RMB’s work on the family Ceratobatrachidae has been supported by the U. S. National Science Foundation (DEB 073199, 0334952, 0743491, 1418895).
#Authors contributed equally to this work
ABSTRACT
In an effort to study the systematic affinities and specieslevel phylogenetic relationships of the enigmatic anurans variably assigned to the genera Ingerana or Limnonectes (family Dicroglossidae), we collected new molecular sequence data for five species including four Himalayan taxa, Limnonectes xizangensis, Lim. medogensis, Lim. alpine, Ingerana borealis and one southeast Asian species, I. tasanae, and analyzed these together with data from previous studies involving other ostensibly related taxa. Our surprising results demonstrate unequivocally that Lim. xizangensis, Lim. medogensis and Lim. alpine form a strongly supported clade, the sister-group of the family Australasian forest frog family Ceratobatrachidae. This discovery requires an expansion of the definition of Ceratobatrachidae and represents the first record of this family in China. These three species are distinguished from the species of Ingerana and Limnonectes by the: (1) absence of interdigital webbing of the foot, (2) absence of terminal discs on fingers and toes, (3) absence of circumarginal grooves on the fingers and toes, and (4) absence of tarsal folds. Given their phylogenetic and morphological distinctiveness, we assign them to the oldest available generic name for this clade, Liurana Dubois 1987, and transfer Liurana from Dicroglossidae to the family Ceratobatrachidae. In contrast, Ingerana tasanae was found to be clustered with strong support with the recently described genus Alcalus (Ceratobatrachidae), a small clade of otherwise Sundaic species; this constitutes a new record of the family Ceratobatrachidae for Myanmar and Thailand. Finally, Ingerana borealis clustered with the “true” Ingerana (family Dicroglossidae), for which the type species is I. tenasserimensis.
Keywords:Dicroglossidae; Himalaya; Liurana
INTRODUCTION1
The frogs of family Ceratobatrachidae (Boulenger, 2009) comprise a morphologically, developmentally, ecologically, and biogeographically greatly variable and, thus, unique clade (Brown et al., 2015). This family is notable for highly variable body size, direct larval development, and the ability to inhabit a wide variety of environments that lackstanding water-from small oceanic islands, to highelevation mossy montane forests (Brown & Alcala, 1982; Brown et al., 2013; Günther, 2015). Currently, 91 species are assigned to three genera: Platymantis Günther, 1858, Cornufer Tschudi, 1838, and Alcalus Brown, Siler, Richards, Diesmos, and Cannatella, 2015 (AmphibiaWeb, 2015; Brown et al., 2015; Frost, 2015). These species are distributed broadly from the South-West Pacific to the island archipelagos of South Asia, with primary centers of species diversity in Philippines and Solomon-Bismarck Archipelago (Brown, 2009; Brown et al., 2013, 2015).
Four species, formerly referred to Southeast Asian frogs Ingerana (Dubois, 1987), were recently assigned to the family Ceratobatrachidae based on molecular data (Brown et al., 2015). The four taxa (I. baluensis, I. mariae, I. rajae, I. sariba) comprise a monophyletic group now shown to be the sister group of Ceratobatrachinae (genera Platymantis and Cornufer). However, “true” Ingerana (based on the phylogenetic position of the type species, Ingerana tenasserimensis [Sclater, 1892]) has been shown in multiple studies to be more closely related to Dicroglossidae (Bossyut et al., 2006; Wiens et al., 2009). Thus, these four species were just recently assigned to the new genus Alcalus in the family Ceratobatrachidae (Brown et al., 2015).
The species in genus Ingerana are small, plump frogs with flattened and expanded toe and finger tips (Dubois, 1987). Thirteen species previously have been referred to this genus on the basis of morphological characters and life history traits. However, recently its members have been placed in different genera, and even different families, based on phylogenetic analysis of molecular data analysis, i.e., A. baluensis, A. mariae, A. rajae and I. tenasserimensis (Bossuyt et al., 2006; Frost et al., 2006; Wiens et al., 2009; Brown et al., 2015). The placement of other Ingerana species was controversial, and some species were tentatively placed in different genera, in the absence of accompanying molecular data. For example, Limnonectes xizangensis was variably assigned to the genera Cornufer (Hu, 1977), Ingerana (subgenus Liurana) (Dubois, 1987), Platymantis (Fei et al., 1990), Micrixalus (Zhao & Adler, 1993), and finally to Limnonectes (subgenus Taylorana) (Borah et al., 2013; Frost, 2015). The complex and convoluted taxonomic placement of several of these species has based on morphological or reproductive characters. Because the few key diagnostic characters emphasized by previous worker are variable, and subject to individual interpretation they may have mislead previous attempts to determine systematic affinities of these poorly known frog species.
Here we report the results of a systematic study of five species variably referred to Limnonectes or Ingerana, including Lim. xizangensis, Lim. medogensis, Lim. alpine, I. borealis and I. tasanae. We redistribute them among two families, according to their phylogenetic affinities, as Liurana xizangensis, Liu. medogensis, Liu. alpine, and Alcalus tasanae (family Ceratobatrachidae) and Ingerana borealis (family Dicroglossidae). These discoveries greatly extend the westernmost geographic distribution of the primarily Australasian archipelago family Ceratobatrachidae into Indochina and China and assign early mainland branching events in this family to lineages now exclusively represented by species with restricted ranges in the high-elevation Himalayan mountains of Tibet.
MATERIALS AND METHODS
Sampling
Four species, Limnonectes xizangensis, Lim. medogensis, Lim. alpine and Ingerana borealis, were sampled from Medog (=Motuo), Tibet (=Xizang), PR China (locality 1 in Figure 1, Table 1). Following the collection of liver tissue samples (preserved in 95% ethanol), the voucher specimens were fixed with 10% formalin and then stored in 70% ethanol. Collection of specimens followed animal-use protocols approved by the Kunming Institute of Zoology Animal Use and Ethics Committee. Two more species, I. tasanae and Occidozyga martensii distributed in Myanmar and Thailand, were also included. We borrowed their tissue samples from the collections of the California Academy of Sciences (CAS), Thailand National History Museum (THNHM), and Field Museum of Natural History (FMNH) (Figure 1, Table 1).
DNA extraction and sequencing
Total DNA was extracted using standard phenol-chloroform protocols (Sambrook et al., 1989). One fragment of mitochondrial DNA of 12S rRNA, tRNA-Val, and 16S rRNA (12S-16S) was sequenced for all samples using primers L2519 and 16Sbr (Table 2). Three partial nuclear DNA sequences of recombination activating gene 1 (Rag1), tyrosinase (Tyr) and rhodopsin (Rhod) were sequenced for all samples using primers included in Table 2. Amplifications were conducted in a 25 uL volume reaction, involved initial denaturing step at 94 °C for 5 min; then 35 cycles of denaturing at 94 °C for 45 sec, annealing at 50 °C or 55 °C for 45 sec, and extending at 72 °C for 45 sec; and a final extending step of 72 °C for 7 min. The products were purified with Gel Extraction Mini Kit (Watson BioTechnologies, Shanghai, China), then sequenced on an ABI 3730×l DNA automated sequencer (Applied Biosystems, UK).
For species not sampled by us, the sequences of 12S-16S, Rag1, Tyr and Rhod were downloaded from GenBank (Table 1). All data were aligned with MUSCLE (Edgar, 2004) and edited using MEGA 5.05 (Tamura et al., 2011).
Phylogenetic analysis
We estimated phylogenetic relationships using Bayesian inference (BI) and maximum parimony (MP) using MrBayes 3.1.2 (Ronquist & Huelsenbeck, 2003) and PAUP* 4.0b10a (Swofford, 2003). Mitochondrial and nuclear sequence data were analyzed separately. Then a phylogenetic tree was conducted using the concatenated sequence of all genes. For BI analysis, the best-fitting nucleotide substitution models were selected for 12S-16S and each codon of Rag1, Tyr and Rhod using the Akaike information criterion in MRMODELTEST v2.3 (Nylander, 2004). The BI analysis used four Markov chains, with default heatingvalues, and run for 5 million generations while sampling trees every 1 000 generations. The first 25% sampled trees were discarded as burn-in, and log-likelihood scores were examined using Tracer v 1.4 (Rambaut & Drummond 2007) to assure convergence (effective sample size [ESS] values >200). For the MP analysis, full heuristic tree searches were used, with 1 000 replications, random addition of sequences and tree-bisection-reconnection (TBR) branch swapping. Non-parametric bootstrap support was estimated using 1 000 replicates of full heuristic searches.
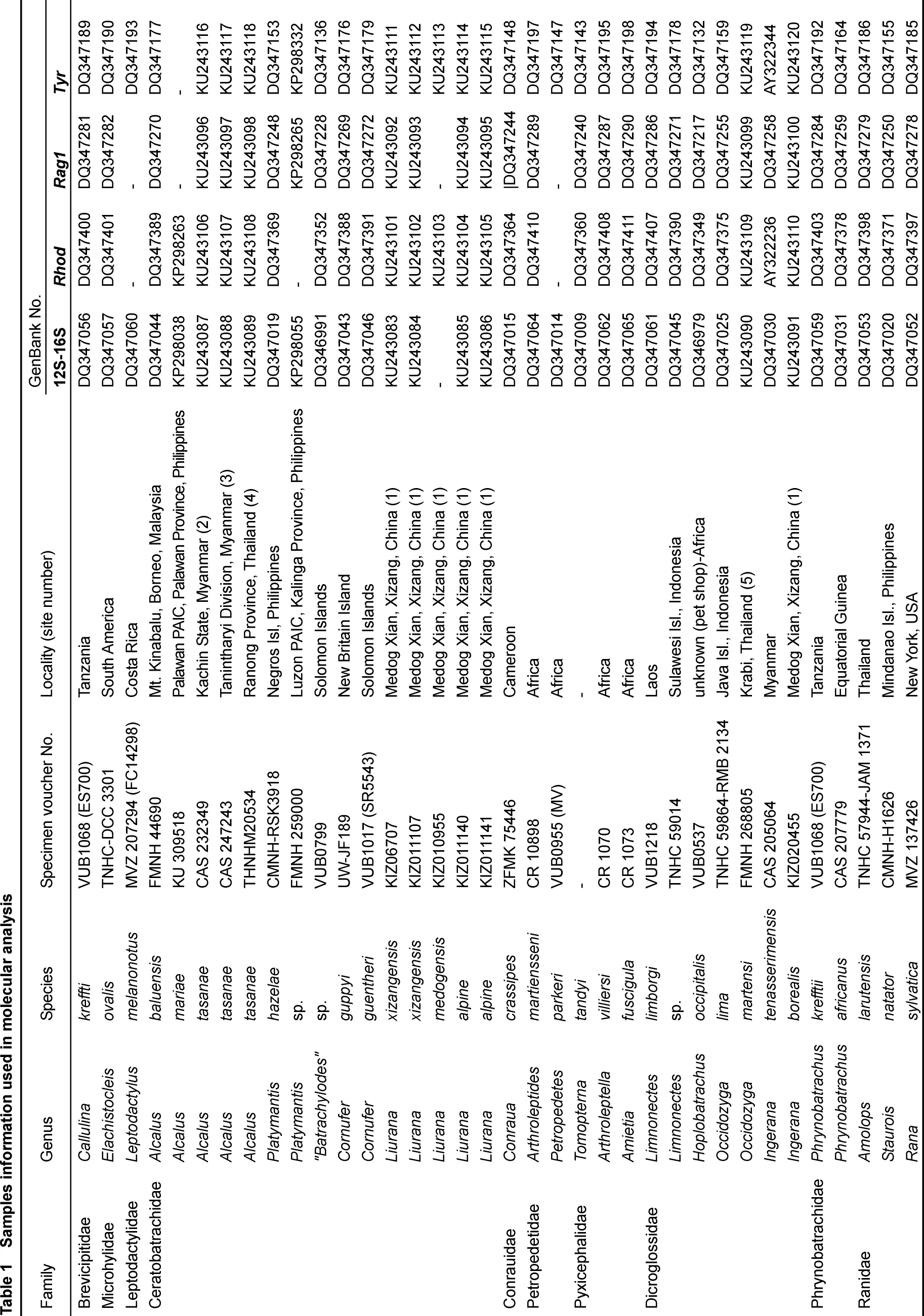

Figure 1 Map of sampling sites
RESULTS
Sequence information
Sequencing generated a total of 1 371 base pairs (bp) of 12S-16S data for Limnonectes alpine and Ingereana tasanae. Additionally, a part of fragment of 12S-16S was successfully sequenced for Occidozyga martensii, Lim. xizangensis and I. borealis. We were unable to collect 12S-16S for Lim. medogensis. For nuclear sequences of Rag1, 1 100 bp was successfully sequenced for all samples except for Lim. medogensis, but we only included 553 bp in subsequent analyses so as to match Rag1 data sequences available on GenBank. Sequences of 553 bp Tyr and 316 bp Rhod were successfully sequenced for all samples. All new generated sequences were submitted to GenBank (Accession numbers KU243083-KU243120, Table 1).

Table 2 Primers information used for four DNA fragments sequencing
Phylogenetic relationships
The best-fitting model were TVM+I+G for mitochondrial 12S-16S, K80+I, TIMef+I and TIMef+I for three codon positions of Rag1, TVM+I+G, K81+I and GTR+G for three codon positions of Tyr, SYM+G, TVM+I+G and TIM+G for three codon positions of Rhod. The phylogenetic analyses based on nuclear DNA and mtDNA showed similar topologies. Most recognized families formed monophyletic groups; however, the monophyly of Dicroglossidae was not recovered using mtDNA, but highly supported by nuclear DNA. This possibly is due to the inability of mtDNA sequence to resolve phylogenetic relationship at deeper levels (i.e., Kingston et al., 2009), or sparse taxon sampling in our analysis. The five focal species were yielded the same topology in both phylogenetic analyses, so the difference between mtDNA and nuclear DNA topologies do not affect our taxonomy. The Bayesian tree resulting from based on concatenated sequence of all genes is shown in Figure 2. Limnonectes xizangensis, Lim. medogensis, Lim. alpine and Ingerana tasanae clustered with species of family Ceratobatrachidae. Three primary lineages were identified in this family, corresponding to twoknown subfamilies Alcalinae (Clade A) and Ceratobatrachinae (Calde B), and a new lineage (Clade C), unsampled in previous phylogenetic estimates (Brown et al., 2015). Samples of Ingerana tasanae from Thailand and Myanmar grouped together, and this clade formed a strongly supported group with Alcalus baluensis and A. mariae (Clade A). Limnonectes xizangensis, Lim. medogensis and Lim. alpine formed a monophyletic group (Clade C), which is strongly supported as related to the family Ceratobatrachidae. Finally, Ingerana borealis samples clustered with species in the subfamily Occidozyginae (Dicroglossidae). This species formed a clade with I. tenasserimensis (type species of Ingerana), as the sister group to Occidozyga.

Figure 2 Bayesian inference tree based on concatenated analysis of all genes
DISCUSSION
Taxonomy of species of Limnonectes and Ingerana, and a record of a new family for China, Myanmar and Thailand
The three poorly understood species, formerly referred to Limnonectes and Ingerana from the largely unexplored area of Himalayan Tibet (Lim. xizangensis, Lim. medogensis and Lim. alpine), have had unstable taxonomic histories (Frost, 2015) and, until now, unclear systematic affinities. Dubois (1987) established the genus Ingerana, in which there are two subgenera Ingerana (Ingerana) and Ingerana (Liurana). Ingerana xizangensis (formerly Cornufer xizangensis, Hu, 1977) was included in subgenus Ingerana (Liurana) by Dubois (1987). Fei et al. (1997) identified significant morphological differences between these two subgenera, including the presence of lingual papilla on the tongue, the absence of terminal discs on fingers and toes, and the absence of circumarginal grooves on fingers and toes in Ingerana (Liurana), (Figures 3-4). Thus, Liurana was elevated to the level of genus to include the species Liu. xizangensis, Liu.medogensis, Liu. alpine and Liu. liui (Fei et al., 1997, 2009, 2012; Huang & Ye, 1997). Fei et al. (2009) considered Liurana to be part of the family Occidozygidae. Subsequently, Fei et al. (2010) established a new subfamily Liuraninae in the family Occidozygidae based on morphological data. Frost et al. (2006) considered Liurana to be a junior synonym of Ingerana on the basis of the original description and overlapping character states. Based on available morphological characters, Borah et al. (2013) placed Liurana in synonymy with Taylorana (now considered to be a subgenus of Limnonectes, [Frost, 2015]). Thus, for the last several years, these species have resided in Limnonectes (Frost, 2015) pending appropriate phylogenetic analysis to determine of their systematic affinities.

Figure 3 Photos of Liurana alpine and Liurana xizangensis in life (Photos by Kai WANG)

Figure 4 Photos of Liurana medogensis (Photos by Kai WANG)
Based on analysis of multilocus DNA sequence data, Liu. xizangensis, Liu. medogensis and Liu. alpine are herein assigned to the family Ceratobatrachidae and represent the first record of this family in China. In our analysis these species formed strongly supported monophyletic group, clustering with members of the Ceratobatrachidae (sensu Brown et al., 2015). In contrast, species of genus Ingerana (I. tenasserimensis and I. borealis) and Limnonectes (Lim. limborgi, Lim. sp.) formed the strongly supported clades in subfamily Occidozyginae, as showed in previous studies (i.e. Bossuyt et al., 2006; Wiens et al., 2009; Pyron & Wiens, 2011). Based on our observations of morphological variation, these three species likewise are distinguished from the species of Ingerana and Limnonectes by the: (1) absence of interdigital webbing of the feet, (2) absence of terminal discs on fingers and toes, (3) absence of circumarginal grooves on the fingers and toes, and (4) absence of tarsal folds. All available evidence supports the recognition of Liu. xizangensis, Liu. medogensis and Liu. alpine as single taxon, for which Liurana is the available generic name with priority. We assign Liurana to the family Ceratobatrachidae. Within Ceratobatrachidae, three lineages are recognized: Clade A and Clade B (Figure 2) correspond to previously recognized subfamilies Alcalinae and Ceratobatrachinae, respectively. The genus Liurana (Clade C) is equivalent in species content to the subfamily Liuraninae Fei, Ye and Jiang, 2010, now transfered to the family Ceratobatrachidae.
Ingerana tasanae is distributed in western and central peninsular Thailand, and its range possibly extends into adjacent Tenaserim and Myanmar (Stuart et al., 2008). Our molecular data clearly place all Ingerana tasanae samples in the same clade as other members of the genus Alcalus (Ceratobatrachidae). This constitutes a new record of family Ceratobatrachidae for Myanmar and Thailand. Our northern Myanmar samples of A. tasanae is highly divergent from individuals from southern Myanmar and southern Thailand. It remains possible that additional taxonomic diversity will be revealed in the genus Alcalus with accumulation of data and field studies of these populations.
Previous studies placed I. borealis in the genus Phrynoglossus (Fei et al., 2009, 2010, 2012), Occidozyga (Ahmed et al., 2009; Mathew & Sen, 2010), and Ingerana (Sailo et al., 2009). Based on our molecular data, I. borealis falls into a strongly supported clade with I. tenasserimensis, the type species of Ingerana. Thus, our molecular data support its systematic position within genus Ingerana based on morphological comparison by Sailo et al. (2009).
New insight from the phylogeny and distribution of Ceratobatrachidae
Brown et al. (2015) developed a stable taxonomy for the family Ceratobatrachidae. Two subfamilies were identified: Ceratobatrachinae and Alcalinae. Ceratobatrachinae includestwo large monophyletic radiations, Cornufer and Platymantis. The species belonging to the subfamily Ceratobatrachinae have a broad distribution in the south-west Pacific, including Philippines, Borneo, New Guinea, Admiralty and Bismarck archipelagos, Solomon Islands, and Fiji. Alcalinae includes only four species of Alcalus, which are distributed only on the island archipelagos of Southeast Asia (Sundaland).
Our research identified other four species which we now transfer to Ceratobatrachidae; this greatly increases the distribution of the family to the mainland of Southeast Asia and the Himalayan region (Figure 1). Given our experience with the unexpected phylogenetic affinities of the species studied here, we would not be surprised if additional phenotypically similar taxa are found to belong in Ceratobatrachidae in the near future. Of particular note, Ingerana charlesdarwini (Das, 1998), distributed in the Andaman Islands (India), could very well be the sister lineage to the remaining lineages in this large and spectacularly diverse anuran family.
The surprising discovery that the clade Ceratobatrachidae is broadly distributed from the Himalayas, mainland and peninsular southeastern Asia, to the southwest Pacific, will help us to understand the biogeography in this region. The sister-group relationship of Ceratobatrachinae and Alcalinae, although not unequivocally supported mirrors the geographic distribution of these clades. This relationship between mainland and archipelago species is also seen in the divergence between the mainland species Alcalus tasanae and the archipelago species A. mariae and A. baluensis. Additional, unexpected patterns between mainland and island taxa may be found with more complete taxon sampling, which emphasizes the need for additional fieldwork in mainland southeastern Asia.
ACKNOWLEDGEMENTS
We would like to thank Dr. Pi-Peng LI (Shenyang Normal University), Mr. Tao LIANG, Mr. Duan YOU, and Mr. Ya-Di HUANG, who helped with fieldwork in Tibet, Prof. Yue-Zhao WANG (CIB), Prof. Yue-Ying CHEN (CIB) and Prof. Sheng-Quan LI (CIB) for kindly letting us examine specimens under their care. We also thank the California Academy of Sciences (CAS), Thailand National History Museum (THNHM), and Field Museum of Natural History (FMNH) for loan of tissue samples.
REFERENCES
Ahmed MF, Das A, Dutta SK. 2009. Amphibians and Reptiles of Northeast India. A Photographic Guide. Guwahati, India: Aaranyak.
AmphibiaWeb: Information on amphibian biology and conservation. [web application]. 2015. Berkeley, California: AmphibiaWeb. Available at http: //amphibiaweb.org/.
Borah MM, Bordoloi S, Purkayastha J, Das M, Dubois A, Ohler A. 2013. Limnonectes (Taylorana) medogensis (Fei, Ye & Huang, 1997) from Arunachal Pradesh (India), and on the identity of some diminutive ranoid frogs (Anura: Dicroglossidae, Occidozygidae). Herpetozoa, 26(1/2): 39-48.
Bossuyt F, Brown RM, Hillis DM, Cannatella DC, Milinkovitch MC. 2006. Phylogeny and biogeography of a cosmopolitan frog radiation: Late cretaceous diversification resulted in continent-scale endemism in the family ranidae.. Systematic Biology, 55(4): 579-594.
Bossuyt F, Milinkovitch MC. 2000. Convergent adaptive radiations in Madagascan and Asian ranid frogs reveal covariation between larval and adult traits. Proceedings of the National Academy of Sciences of the United States of America, 97(12): 6585-6590.
Boulenger GA. 2009. Diagnoses of new reptiles and batrachians from the Solomon Islands, collected and presented to the British Museum by Guppy HB, Esq, MB, ‘Lark’ HMS. Proceedings of the Zoological Society of London, 52(2): 210-213.
Brown WC, Alcala AC. 1982. Modes of reproduction of Philippine anurans. In: Rodin AGJ, Miyata A., (eds). Advances in Herpetology and Evolutionary Biology. Cambridge, MA: Museum of Comparative Biology, 416-428.
Brown RM. 2009. Frogs. In: Gillespie R, Clague D. (, eds). Encyclopedia of Islands. Berkeley: University of California Press, .
Brown MR, Richards JS, Broadhead ST. 2013. A new shrub frog in the genus Platymantis (Ceratobatrachidae) from the Nakanai Mountains of eastern New Britain Island, Bismarck Archipelago. Zootaxa, 3710 (1): 31-45. Brown MR, Siler DC, Richards JS, Diesmos AC, Cannatella CD. 2015. Multilocus phylogeny and a new classification for Southeast Asian and Melanesian forest frogs (family Ceratobatrachidae). Zoological Journal of the Linnean Society, 174(1): 130-168.
Das I. 1998. A remarkable new species of ranid (Anura: Ranidae), with phytotelmonous larvae, from Mount Harriet, Andaman Island. Hamadryad, 23: 41-49.
Dubois A. 1987. Miscellanea taxinomica batrachologica (I). Alytes, 5: 7-95. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32(5): 1792-1797.
Fei L, Ye CY, Huang YZ. 1990. Key to Chinese Amphibians. Chongqing: Publishing House for Scientific and Technological Literature.
Fei L, Hu SQ, Ye CY, Huang YZ. 2009. Fauna Sinica. Amphibia. Volume 3. Anura Ranidae. Beijing: Science Press.
Fei L, Ye CY, Huang YZ. 1997. Taxonomic studies of the genus Liurana of China including descriptions of a new species (Amphibia: Ranidae). Cultum Herpetologica Sinica, 6-7: 75-80.
Fei L, Ye CY, Jiang JP. 2010. Phylogenetic systematics of Ranidae. Herpetologica Sinica, 12: 1-43.
Fei L, Ye CY, Jiang JP. 2012. Colored Atlas of Chinese Amphibians and Their Distributions. Chengdu: Sichuan Publishing House of Science & Technology, . .
Frost RD, Grant T, Faivovich J, Bain HR, Haas A, Haddad FBC, de Sá OR, Channing A, Wilkinson A, Donnellan CS, Raxworthy JC, Campbell AJ, Blotto LB, Moler P, Drewes CR, Nussbaum AR, Lynch DJ, Green MD, Wheeler CW. 2006. Amphibians tree of life. Bulletin of American Museum of Natural History, 297: 1-370.
Frost DR. 2015. Amphibian Species of the World: an Online Reference. Version 6.0 (Nov 12, 2015). Electronic Database accessible at http: // research.amnh.org/herpetology/amphibia/index.html. American Museum of Natural History, New York, USA.
Günther, ACLG. 1858. Neue Batrachier in der Sammlung des britischen Museums. Archiv für Naturgeschichte, 24: 319-328.
Günther R. 2015. Description of two new taxa of the ceratobatrachid genus Platymantis from western New Guinea (Amphibia, Anura). VertebrateZoology, 65(1): 101-116.
Hu SQ. 1977. A survey of amphibians in Xizang (Tibet). Acta Zoologica Sinica, 23: 54-63.
Huang YZ, Ye CY. 1997. A new species of the genus Liurana (Amphibia: Ranidae) from Xizang, China. Cultum Herpetologica Sinica, 6-7: 112-115. Kingston SE, Adams LD, Rosel PE. 2009. Testing mitochondrial sequences and anonymous nuclear markers for phylogeny reconstruction in a rapidly radiating group: molecular systematics of the Delphininae (Cetacea: Odontoceti: Delphinidae). BMC Evolutionary Biology, 9: 245.
Mathew R, Sen N. 2010. Pictorial Guide to Amphibians of North East India. Kolkata, India: Zoological Survey of India.
Nylander JAA. 2004. MrModeltest v2. Uppsala University, Evolutionary Biology Center.
Palumbi SR, Martin A, McMillan WO, Stice L, Grabowski G. 1991. The simple fool’s guide to PCR, Version 2.0. Privately published document compiled by S. Palumbi.
Pyron RA, Wiens JJ. 2011. A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of advanced frogs, salamanders, and caecilians. Molecular Phylogenetics and Evolution, 61(2): 543-583.
Rambaut A, Drummond AJ 2007. Tracer v 1.4. Available at http://beast.bio.ed.ac.uk.sci-hub.io/Tracer.
Richards CM, Moore WS. 1996. A phylogeny for the African treefrog family Hyperoliidae based on mitochondrial rDNA. Molecular Phylogenetics and Evolotion, 5(3): 522–532.
Ronquist F, Huelsenbeck JP. 2003 MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19(12): 1572-1574.
Sailo S, Lalremsanga HT, Hooroo RNK, Ohler A. 2009. Ingerana borealis (Annandale, 1912): a new record from Mizoram (India), with notes on its systematic position and natural history. Alytes, 27: 1-12.
Sambrook J, Fritsch E, Maniatis T. 1989. Molecular Cloning: A Laboratory Manual (2nd edn). New York: Cold Spring Harbor Laboratory Press, .
Stuart BL. 2008. The phylogenetic problem of Huia (Amphibia: Ranidae). Molecular Phylogenetics and Evolution, 46(1): 49-60.
Stuart SN, Hoffmann M, Chanson J, Berridge NCR, Ramani P, Young B. 2008. Threatened Amphibians of the World. Barcelona, Spain; International Union for the Conservation of Nature, Gland. Switzerland; Conservation International, Arlington, Virginia, USA: Lynx Editions.
Swofford DL. 2003. PAUP*: Phylogenetic analysis using parsimony (* and other methods) Version 40b10. Sinauer Associates, Sunderland, Maryland.
Tamura K, Peterson D, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28(10): 2731-2739.
Tschudi JJ. 1838. Classification der Batrachier mit Berücksichtigung der fossilen Thiere dieser Abtheilung der Reptilien. Neuchâtel: Petitpierre.
Wiens JJ, Sukumaran J, Pyron RA, Brown RM. 2009. Evolutionary and biogeographic origins of high tropical diversity in Old World frogs (Ranidae). Evolution, 63(5): 1217-1231.
Zhao EM, Adler KA. 1993. Herpetology of China.Oxford, Ohio: Contributions to Herpetology 10. Society for the Study of Amphibians and Reptiles.
DOI:10.13918/j.issn.2095-8137.2016.1.7
*Corresponding author, E-mail: chej@mail.kiz.ac.cn
杂志排行
Zoological Research的其它文章
- New Year address from Zoological Research
- Advances in herpetological research emanating from China
- A new genus and species of treefrog from Medog, southeastern Tibet, China (Anura, Rhacophoridae)
- A new species of the genus Scutiger (Anura: Megophryidae) from Medog of southeastern Tibet, China
- A new species of the genus Amolops (Amphibia: Ranidae) from southeastern Tibet, China
- Two new species of Japalura (Squamata: Agamidae) from the Hengduan Mountain Range, China
