Isolation and Identification of Nitrite-oxidizing Bacteria
2015-02-06YulongCHENWenyongTANDaYANG
Yulong CHEN,Wenyong TAN,Da YANG
1.Sichuan Preschool Educators College,Jiangyou 621709,China;2.Genetic Engineering Research Centre,School of Life Science,Chongqing University,Chongqing 400030,China
1 Introduction
With the rapid development of China's high-density aquaculture,the nitrite accumulation in the water is becoming very prominent.Excessive nitrite content can not only cause a variety of metabolism disorders but also directly lead to aquaculture animal diseases or death,so nitrite poses a serious threat to farming and intensive aquaculture[10,24].Therefore,the prevention of nitrite accumulation in water has become one of the key measures to promote sustainable development of intensive aquaculture.Currently we can use physical,chemical and biological methods to degrade nitrite.Noticeably,using chemical method for nitrite degradation generates little success,and it easily leads to a series of environmental and social issues.The use of microbial agents for nitrite degradation will cause low costs and infinitesimal environmental pollution,so it is a good way to restore the environment,and reduce nitrite concentrations in aquaculture waters[20].For example,nitrobacteria andBacillus subtilis,etc.have achieved remarkable results in many countries.Based on this,we take strains with nitrite degradation capacity from the shrimp farming pond silt in Hepu City,and after screening and physiological,chemical and 16S rRNA identification,it is identified asPannonibacter phragmitetus,which lays the foundation for further study.
2 Materials and methods
2.1 Experimental materialsAquaculture silt is collected from shrimp farming pond in Hepu City.Nitrite-oxidizing bacteria selective medium:Na2HPO412H2O;5 g/L,KH2PO4;1 g/L,MgSO47H2O;0.1g/L,sodium succinate 6 g/L,NaNO22 g/L,agar 2 g/L;trace element solution 2 mL;pH=8.Trace element solution(g/L):EDTA 50 g/L;ZnSO42.2 g/L;CaCl25.5 g/L;MnCl24H2O 5.6 g/L;FeSO47H2O 5 g/L;(NH4)6Mo7O24H2O 1.1 g/L;CuSO45H2O 1.6 g/L;CoCl26H2O 1.6 g/L;pH=7.Nacl solution:0.9%.The above culture medium and solutions are sterilized at121℃ for 20min,and stored in the refrigerator.PCR primers are synthesized by Shanghai Sangon Company;Taq polymerase,PCR buffer,Mg2+and dNTPsare provided by Takara Company;the gel extraction kit is provided by Shanghai Sangon Company;other reagents are analytically pure reagents produced at home.
2.2 Isolation,purification,screening and biochemical identification of nitrite-oxidizing bacteria10g of the soil sample is dissolved in 100ml of sterile 0.9%Nacl solution and kept stationary at 28℃ after overnight incubation.0.9%Nacl solution is used to dilute the culture solutions 10-108fold,and the culture solutions of different dilution rates are evenly coated on the denitrifying bacteria culture plate for the separation and purification of n Nitrite-oxidizing bacteria.
2.3 Molecular identification of nitrite-oxidizing bacteriaThe 16S rRNA sequence identification method for strain HP007 is as follows:the activated HP007 is inoculated on denitrifying bacteria medium plate,and the single colonies are picked directly for PCR amplification.The bacterial universal primers are used for PCR amplification[9],and the sequence is as follows:Primer 1(F27):5′-AGAGTTTGATCATGGCTCAG-3′;Primer 2(R1492)5′-TACGGTTACCTTGTTACGACTT-3′.PCR reaction system:10×Buffer 2.5μl,MgCl22μl(25 mmol/L),dNTPs Mixture(10 mmol/L)0.5μl each,Primer 1 and Primer 2(10μmol/L)1μl each,DNA Polymerase(5u.μ/L)0.2μl,Template 1μl,dd H2O 16.8μl,total volume of 25μl.PCR reaction conditions:94℃ 5 min;94℃ 30 s;50℃ 1 min;72℃ 2 min,30 cycles;72℃ 10min.After completion of the reaction,5μl of the reaction solution is taken for electrophoresis using 1%agarose gel.The PCR products are purified by DNA gel extraction kit,and it is commissioned by Shanghai Sangon Company.After using DNAMAN software for editing the identification results,the BLAST software and the known 16S rRNA sequence in GenBank database are used for homology comparison to select the sequences with high homology.After multiple comparisons using BioEdit 7.0 and MEGA 6.0,the neighbor-joining method is used to build the phylogenetic tree.The strain type is determined based on colonial morphology and physiological and biochemical characteristics.
3 Results
3.1 The morphological character and physiological and biochemical identification of strainAfter 20 silt samples are diluted and screened,we get a strain with ability to degrade nitrite.This strain is cultured on solid nitrite culture medium for15 d(28℃),and the colony diameter is0.8-1.0mm(Colonieson alkaline agar medium are whitish-beige,circular,convex,smooth and shiny with entire edges.Cells are non-spore-forming rods,motile with polar flagella,contain PHA and stain Gram-negative,and facultatively anaerobic,chemo-organotrophic.)(Fig.1).The physiological and biochemical characteristics are shown in Table 1.Based on Bergey Bacterial Identification Manual9[9,11]and literature[2],it is found that the strain is in line with the Pannonibacter phragmitetus features.
3.2 Determination and analysis of 16S rDNA sequence of HP007 strainUsing the bacterial 16S rDNA universal primers for single colony PCR amplification,we get 1.5 kb of DNA fragment from HP007 strain(Fig.2),and directly send the PCR products to Shanghai Sangon Company for bi-directional DNA sequencing.By comparing the sequencing results with BLAST of GenBank,it is found that the sequence homology between 16S rDNA sequence of HP007 strain andPannonibacter phragmitetusstrain C6-19 reaches 99% (Fig.3).Therefore,based on homology of 16S rDNA sequence,combined with comprehensive analysis of mycelial morphology and physiological and biochemical characteristics of HP007 strain,this strain is identified asPannonibacter phragmitetus.According to the screening location,it is named HPPP007 strain.
4 Conclusions and discussions
In order to select the strain that can degrade nitrite,we use the screening plate with nitrite as the sole nitrogen source to select the strain with ability to degrade nitrite,and get a strain with nitrite degrading capacity from the silt of shrimp farming pond in Hepu City,Guangxi Zhuang Autonomous Region.By identifying the strain from colony morphology,physiological and biochemical characteristics and 16S rRNA sequence,we finally get a bacteria strain that can degrade nitrite,and this strain can grow well on the culture medium with nitrite concentration of2 g/L.Based on morphology,nitrogen source requirements and evolutionary tree analysis of the above 16S rRNA sequence,it is found that this strain belongs toPannonibacter phragmitetus,which is completely different from the previously reportedNitrobacter,Ni-trococcus,Nitrospina,andNitrospira[15-18].This study is a complement to nitriteoxidizing bacteria flora.Pannonibacter phragmitetusas a new spe-cies was reported in 2003[2],and in recent years,its efficient Cr(Ⅵ)deoxidization function has been found[4].There is a need to conduct further experiments on whether we can use Pannonibacter phragmitetus as the microbial agent to reduce nitrite damage in farming and repair environment.
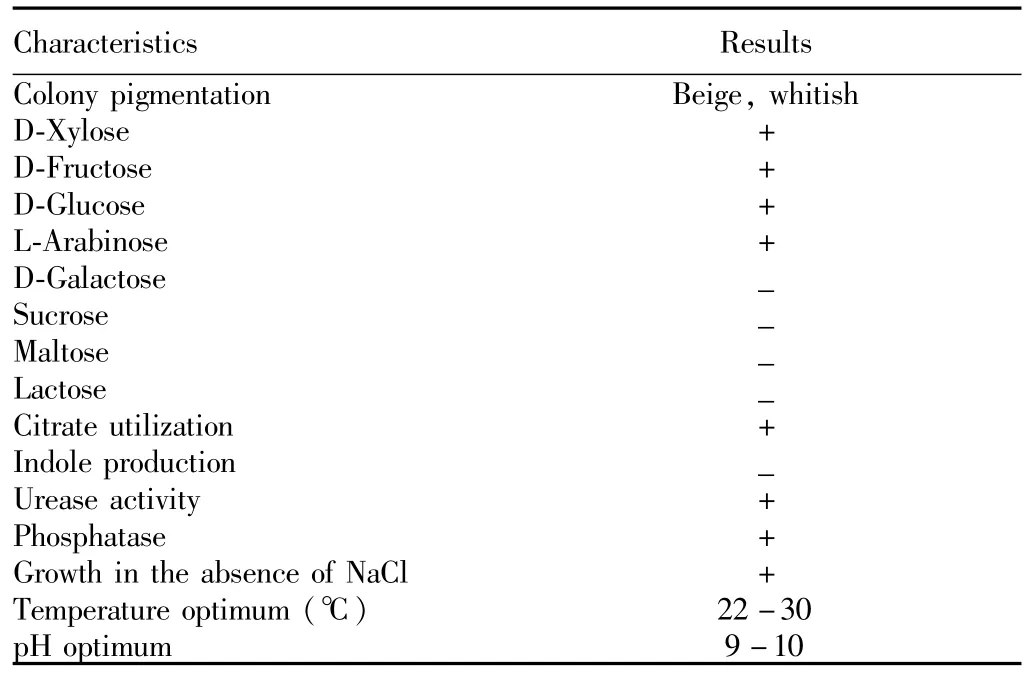
Table 1 Physiological and biochem ical characteristics of HP007 strain
[1]Anzai Y,KimH,Park JY,et al.Phylogenetic affiliation of the pseudomonads based on16S rRNA sequence[J].Int JS yst Evol Microbiol,2000,50(4):1563-1589.
[2]BorsodiAK,Micsinai A,Kovacs G,etal.Pannonibacter phragmitetusgen.nov.,sp.nov.,a novel alkalitolerant bacterium isolated from decomposing reed rhizomes in a Hungarian soda lake[J].Int J Syst Evol Microbiol,2003(53):555-561.
[3]Buchanan RE,Gibbons NE.Bergey's Manual of Determinative Bacteriology[M].Beijing China Science Press,1989.
[4]CHAILy,XU Yz,WANGHy,etal.Cr(Ⅵ)remediation by Pannonibacter phragmitetus in contaminated soils[J].The Chinese Journal of Nonferrous Metals,2009(12):2230-2236.
[5]CHEN F,XIA Q,JU LK.Aerobic denitrification of Pseudomonas aeruginosa monitored by online NAD(P)H fluorescence[J].Appl Environ Microbiol,2003,69(11):6715-6722.
[6]Chen HJ,Huang SL,Tseng DH.Aerobic biotrans formation of octylphenol polyethoxylate surfactant in soil microcosms[J].Environ Technol,2004,25(2):201-210.
[7]Clarridge JEó.Impact of 16SrRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases[J].Clin Microbiol.Rev,2004,17(4):840-862.
[8]Drancourt M,Bollet C,Carlioz R,et al.16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates[J].J Clin Microbiol,2000,38(10):3623-3630.
[9]Dong XZ,CAIMY.Manual of Systematic and Determinative Bacteriology[M].Beijing,China:Science Press,2001.
[10]David JR,Nicholas JW.Inorganic nitrogenme-tabolism in bacteria[J].Current Opinion in Chemical Biology,1999(3):207-219.
[11]FAN XR,SHEN P.Experiment of Microbiology[M].Beijing,China:The People’s Education Press,1989:130-131.
[12]Patureau D,Bernet N,Delgenes JP,etal.Effect of dissolved oxygen and carbon-nitrogen loads on denitrification by an aerobic consortium[J].Appl Microbiol Biotechnol,2000,54(4):535-542.
[13]Patureau D,Godon JJ,Dabert P,etal.Microvirgula aerodenitrificansgen.nov.,sp.nov.,a new gram-negative bacterium exhibiting co-respiration of oxygen and nitrogen oxidesup to oxygen-saturated conditions[J].Int J Syst Bacteriol,1998,48(3):775-782.
[14]Patureau D,Helloin E,Rustrian E,etal.Combined phosphate and nitrogen removal in a sequencing batch reactor using theaerobic denitrifier,Microvirgula aerodenitrificans[J].Water Res,2001,35(1):189-197.
[15]Patureau D,Zumstein E,Delgenes JP,et al.Aerobic denitrifiers isolated from diverse natural and managed ecosystems[J].Microb Ecol,2000,39(2):145-152.
[16]Robertson L A,Kuenen JG.Aerobic denitrification:a controversy revived[J].Arch Microbiol,1984(139):351-354.
[17]Takaya N,Catalan-SakairiMA,Sakaguchi Y,et al.Aerobic denitrifying bacteria that produce low levels of nitrous oxide[J].Appl Environ Microbiol,2003,69(6):3152-3157.
[18]Ka JO,Urbance J,Ye RW,etal.Diversity of oxygen and N-oxide regulation of nitrite reductases in denitrifying bacteria[J].FEMS Microbiol Letter,1997,156(1):55-60.
[19]Vollack KU,Hartig E,KornerH,etal.Multiple transcription factorsof the FNR family in denitrifying Pseudomonas stutzeri:characterization of four fnr-like genes,regulatory responses and cognate metabolic processes[J].Mol Microbiol,1999,31(6):1681-1694.
[20]Su JJ,Liu BY,Lin J,etal.Isolation of an aerobic denitrifying bacterial strain NS-2 fromthe activated sludge of piggery waste water treatment systems in Taiwan possessing denitrification under 92%oxygen atmosphere[J].JAppl Microbiol,2001,91(5):853-860.
[21]OtaniY,Hasegawa K,HanakiK.Comparison of aerobic denitrifying activity among three cultural species with various carbon sources[J].Water Sci Technol,2004,50(8):15-22.
[22]Ozeki S,Baba I,Takaya N,et al.A novel C1-using denitrifier alcaligenes sp.STC1 and its genes for copper-containing nitrite reductase and azurin[J].Biosci Biotechnol Biochem,2001,65(5):1206-1210.
[23]Kong Q X,Li JW,WangXW,et al.Anewscreening method for aerobic denitrification bacteria and isolation of a novel strain[J].Ch in JAppl Environ Biol,2005,11(2):222-225.
[24]Lin JT,Stewart V.Nitrate assimilation by bacteria[J].AdvMicrob Physio,1998(39):330-379.
[25]Romano G,Stampi S,Zanetti F,et al.Occurrence of gram-negative bacteria in drinkingwater undergoing softening treatment[J].Zentralbl Hyg Umweltmed,1997,200(2-3):152-162.
[26]Guo QY,Xu Z,Yang X S.Bacterial diversity in cultured Pseudosciaenacrocea from the East China Sea[J].Marine Fisheries,2005,27(3):241-245.
[27]Ivanov V,Stabnikov V,ZhuangW Q,et al.Phosphate removal from the returned liquor of municipal wastewater treatment plant using iron-reducing bacteria[J].JAppl Microbiol,2005,98(5):1152-1161.
[28]Park M,Kim C,Yang J,et al.Isolation and characterization of diazotrophic growth promotingbacteria from rhizosphere of agricultural crops of Korea[J].Microbiol Res,2005,160(2):127-133.
杂志排行
Asian Agricultural Research的其它文章
- Empirical Study on Formation Path of Fam ily Farms in Different Environments from the Perspective of ANT
- An Empirical Analysis of the Factors Influencing the Cooperative Relationship between Broiler Processing Enterprises and Broiler Raisers
- Prediction of Coal Consumption in China Based on the Partial Linear M odel
- How to Develop Rubber Production in Xishuangbanna Dai Autonomous Prefecture?
- Relation between Rural Financial Development and Rural Economic Grow th:A Case Study of Yunnan Province
- A Study of Regional Brand Positioning Strategy for Linhai Orange Based on Soft Laddering
