Genetic Inheritance and Molecular Marker of Clubroot Resistance Genes in Brassica campestris ssp.chinensis
2015-02-05BoSONGHuanSUOLongzhengCHENHaiXUXiaoxueFANHuiZHANGXihanYUAN
Bo SONG,Huan SUO,Longzheng CHEN,Hai XU,Xiaoxue FAN,Hui ZHANG,Xihan YUAN
Institute of Vegetable Crops,Jiangsu Academy of Agricultural Sciences,Nanjing 210014,China
Genetic Inheritance and Molecular Marker of Clubroot Resistance Genes in Brassica campestris ssp.chinensis
Bo SONG,Huan SUO,Longzheng CHEN,Hai XU,Xiaoxue FAN,Hui ZHANG,Xihan YUAN*
Institute of Vegetable Crops,Jiangsu Academy of Agricultural Sciences,Nanjing 210014,China
[Objective]This study was conducted to investigate the genetic inheritance of clubroot resistance in Chinese non-heading cabbage(Brassica campestrisssp.chinensis).[Method]The clubroot resistance gene was introduced from aBrassica campestrisssp.pekinensiscultivar to non-heading Chinese cabbage,and the inheritance and molecular markers of clubroot resistance gene in parental lines,F1,F2and BC1of non-heading Chinese cabbage were studied through pathogen inoculation at seedling stage and ISSR-PCR.[Result]Clubroot resistance in non-heading Chinese cabbage was controlled by a single dominant gene.ISSR molecular markers with Bulk segregant analysis(BSA)found that primer-873 was linked to resistance gene,named CR-873,and the genetic distance between the marker and the resistance gene was 9.72 cM.[Conclusion]The results provide references for the molecular marker assisted breeding of non-heading Chinese cabbage.
Brassica campestrisssp.chinensis;Clubroot resistance;Molecular marker
N on-heading Chinese cabbage (Brassica campestrisssp.chinensis)which is native to China,is one of the important green vegetables and widely planted in Chi na,especially in south China[1].Clubroot,a worldwide disease caused byPlasmodiophora brassicaeWoronin, can influence the nutrient absorption and transfer in roots and stems of cruciferous crops,leading to poor growth, wilting,short plants,poor quality and yield reduction,and finally resulting in huge economic losses[2-3].In recent years,soil-borne diseases have become an important factor limiting the development of non-heading Chinese cabbage.Cubroot gradually emerges and spreads when cabbage is planted repeatedly.
So far,studies on molecular markers linked to clubroot resistance gene have been performed mainly onBrassica campestrisandBrassica oleracea.In these studies quantitative trait locus(QTL)related to clubroot resistance were identified using RAPD and RFLP[4].Gene locus related to clubroot resistance in heading Chinese cabbage(Brassica campestrisssp.pekinensis)has been mapped.It was also reported that the genetic inheritance of clubroot resistance varied among different physiological races of cabbage[5].
Siet al.[6]found that clubroot resistance was controlled by above three pairs of genes and incompletely recessive inB.oleracea.However, some other studies showed that clubroot resistance inB.napuswas controlled by 1-2 independent dominant genes,and these genes can be found in most clubroot-resistant plants[7-9].
Non-heading Chinese cabbage clubroot is a soil-borne disease and difficult to control,and has been rarely reported so far.Therefore,a specific molecular marker for clubroot resistance is important to improve breeding efficiency and conduct marker-assisted breeding.In this study,hybridization between a clubroot-resistant Brassica campestris ssp.pekinensis cultivar and non-heading Chinese cabbage was carried out,and the clubroot resistance of the parents,F1,F2and BC1generations was identified to screen the molecular marker linked to clubroot resistance gene,and the results provide references for the molecular marker assisted breeding of non-heading Chinese cabbage.
Materials and Methods
Materials
A Brassica campestris ssp. pekinensis cultivar highly resistant to clubroot(CR)was introduced from the National Horticultural Research Institute of Korea in 2010.Then,hybridization between this CR material as the female parent,and T Qing which was provided by Institute of Vegetable Crops,Jiangsu Academy of Agricultural Sciences was carried out to obtain their F1generation.In the spring of 2011,the parental materials and their F1generation were planted in the experimental plots of the Institute of Vegetable Crops,Jiangsu Academy of Agricultural Sciences.F2generation was obtained by selfing F1generation, and BC1generation was obtained by backcrossing F1with the female parent material CR.Conventional field managements were carried out.
Methods
Resistance identification of all the generationsThree repetitions for each of T Qing,CR,F1,F2and BC1generations were planted in 50-hole trays and randomly arranged,with thirty plants in each repetition.
The seedlings at three-leaf stage were transplanted into pots to identify their resistance to clubroot.Briefly,1 ml of spore suspension containing 5× 107spores was dropped to the rhizosphere of host plants.Six weeks later, the roots of the plants were dig out and washed to investigate clubroot incidence and to calculate disease index. Field disease incidence was classified into five levels based on previous studies with slight modification[3].In detail,level 0:clubroot disease did not occur;level 1:a few small tumors were found on fibrous root and lateral roots but not found on the main root; level 2:most tumors were found on fibrous roots,more on lateral roots,and a few on the main root;level 3:the main root was severely infected and swollen,and most fibrous and lateral root had tumors;level 4:the main root was very swollen,with very few fibrous roots.Plants of level 0 were resistant to clubroot,and plants of other level were susceptible to clubroot.
DNA extraction and detection
DNA was extracted from young leaves of cabbage using CTAB method.The integrity and purity of DNA sample extracted were detected through electrophoresis on 1.0%agarose gel,and its quality and concentration were measured using a UV spectrophotometer.Finally the DNA solution was diluted to 15 ng/μl for later analysis.
Construction of clubroot-resistant and susceptible gene pools and screening of primersBulk segregant analysis(BSA)was performed for clubroot resistance gene mapping. Equal volumes of the DNA samples of 10 randomly selected clubroot-resistant seedlings and 10 randomly selected clubroot-susceptible seedlings of F2population were separately bulked to establish the clubroot-resistant and susceptible gene pools.Then, ISSR primers those generated specific bands between the two pools were selected.
ISSR-PCRThe ISSR-PCR system (20 μl)contained 1×PCR buffer,30 ng of template DNA,1 U of Taq polymerase,250μmol/L dNTPs,0.25 μmol/L each primer and 2.5 mmol/L Mg2+.The reaction was started with predenaturing at 94℃for 5 min,followed by 35 cycles of denaturing at 94℃for 1 min,annealing at 52℃for 1 min,extension at 72℃for 90 s,and ended with a final extension at 72℃for 10 min.
Agarose gel electrophoresisPCR products were separated through electrophoresis on 2.0%agarose gel containing 0.5 μg/ml EB,with DNA MarkerⅤ(Tiangen)as the molecular weight standard,and visualized under a UV imaging system.
ISSR marker linkage analysisThe screened ISSR markers were tested between the parental lines and F2segregating population,and detected through electrophoresis on 2.0% agarose gel.The target bands were found in clubroot-resistant parent,and not found in clubroot-susceptible parent.F1generation was susceptible to clubroot and had the target bands. Among F2population,the recombinant individuals were resistant to clubroot and did not have the target bands;the heterozygous individuals were susceptible to clubroot and did not had the target bands;and the homozygous individuals were susceptible to clubroot and had the target bands.Finally,the gene exchange rate was calculated from the number of recombinant individuals in F1population,and then converted to Morgan genetic distance according to Kosambi’s function for gene mapping.
Results and Analysis
Identification of clubroot resistance
As shown in Table 1,all the 20 infected seedlings of parent CR were highly resistant to clubroot and identified to be level 0 according to their clubroot incidence,while all the 20 infected seedlings of parent T Qing were highly susceptible to clubroot and identified to be level 3 or above according to their clubroot incidence.All the 30 infected seedlings of F1generation were highly resistant to clubroot and identified to be level 0 according to their clubroot incidence.Among the 73 infected seedlings of F2generation randomly selected,54 were highly resistant(level 0),and 19 were susceptible to clubroot(one in level 1,two in level 2 and 16 in level 3).So,the ratio of clubroot-resistant seedlings to clubroot-susceptible seedlings of F2generation was 54:19,approximately equaling to 3:1(χ2=0.074<X20.05= 3.84).Among the 70 randomly selected infected seedlings of BC1 generation,36 were highly resistant(level 0), and 34 were susceptible to clubroot (two in level 1,one in level 2 and 31 in level 3).So,the ratio of clubroot-resistant seedlings to clubroot-susceptible seedlings of BC1generation was 36:34,approximately equaling to 1:1 (χ2=0.057<X20.05=3.84).In summary, no seedling of level 4 was found,indicating that the clubroot resistance was controlled by a single dominant gene in non-heading Chinese cabbage.
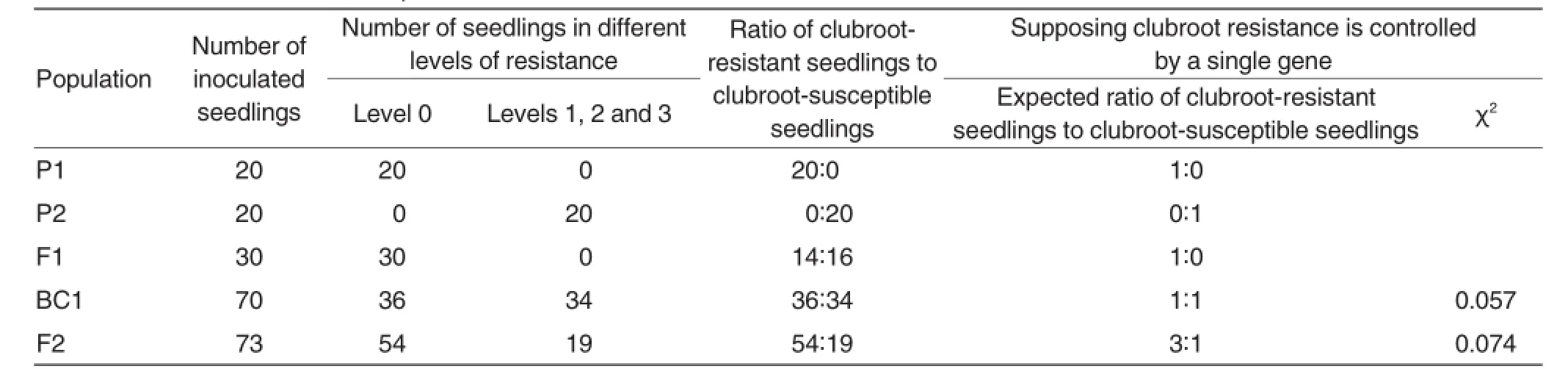
Table 1Resistance to clubroot in parental lines,F1,F2and BC1
Molecular marker linked to clubroot resistance gene in non-heading Chinese cabbage
Among the 90 ISSR primers used for PCR amplification of parent lines,21 generated clear and stable polymorphic band between the parents,accounting for 23.0%of total. Then,the 21 primers were screened between club-resistant and susceptible gene pools and the results turned out that primer 873(5’-GACAGACAGACAGAC A-3’)could amplify a polymorphic band about 500 bp (Fig.1).
As the clubroot resistance gene is dominant,the polymorphic band can be amplified from homozygous and heterozygous individuals resistant to clubroot but can not be amplified from homozygous individuals susceptible to clubroot if the ISSR marker and the clubroot resistance gene were closely linked.So,primer 873 was further verified using 73 individuals of F2generation.It was turned out that among the 54 highly clubroot-resistant individuals (with resistance of level 0)primer 873 amplified the polymorphic band from 53 individuals,and did not from the last one.Among the 19 individuals susceptible to clubroot,primer 873 amplified the polymorphic band from six individuals(Fig.2).In summary,the marker 873 and the resistance gene recombined in seven individuals,so the gene exchange rate was 9.59%.According to Kogambi’s mapping function,the genetic distance between the marker named CR-873 and the resistance gene was 9.72 cM.
Discussion
The genetic inheritance of clubroot resistance gene was mainly studied in three Brassica crops B.campestris,B.oleracea and B.napus. Laurens et al.[10]believed that clubroot resistance in B.oleracea was controlled by several alleles which were incompletely dominant,and had significant additive genetic effects.Voorrips et al.[11]clubroot resistance was controlled by recessive genes in B.oleracea.At least three clubroot resistance genes were found in B.rapa, and these genes were relatively independent,and played different roles in the physiological races of P.brassicae[12-14].Diederichsen et al.[15]found that clubroot resistance was controlled by a major gene and at least two recessive genes in B.napus.The study of Wang et al.[16]revealed that most resistance genes of B.rapa were introduced from Europe forage turnip;one or several resistance genes were obtained by different varieties and these genes were relatively independent. Our results showed that the clubroot resistance was controlled by a single dominant gene in non-heading Chinese cabbage.
Matsumoto et al.[17]found that two RFLP markers were linked to a CR gene CRa,located them on chromosome R3 with a genetic distance of 34 cM[17].Suwabe et al.[18]found two CR loci Crr1 and Crr2 using SSR markers. Using the Siloga as the CR source, Kuginuki et al.[19]found that three RAPD markers were linked to CR locus.ISSR marker can reveal more genome polymorphism than RFLP, SSR and RAPD,and it is more stable than RAPD and simpler to operate than AFLP.In this study,BSA and ISSR were used in combination to screen the gene markers linked to clubroot resistance gene in non-heading Chinese cabbage,and one molecular marker which was closely linked to clubroot resistance gene was obtained, named CR-873.The genetic distance between the marker and the resistance gene was 9.72 cM.The genetic distance was relatively large,probably because the population used to calculate genetic distance was smaller which had some influence to the calculation.In future work,we plan to expand the population for calculating the genetic distance and to convert theISSR markers linked to clubroot resistance gene to SCAR maker which is more stable.
[1]Ministry of Agriculture of the People’s Republic of China(农业部).Vegetable planting area and yield in the provinces/ cities of China in 2006(2006年全国各地蔬菜播种面积和产量)[J].China Vegetables(中国蔬菜),2008,3(1):65.
[2]LIU Y(刘勇),YANG WQ(杨文强), HUANG XQ(黄小琴),et al.Main factors affecting the germination of dormant spores of Plasmodiophora brassicae, which can cause clubroot to Chinese cabbage(影响小白菜根肿病菌休眠孢子萌发的主要因子研究)[C]//Proceedings of the annual meeting of Chinese society for fungi in 2009(中国菌物学会2009学术年会论文摘要集).Beijing Science Press(北京:科学出版社),2009.
[3]YANG WQ(杨文强).Study on the occurrence regulation,pathogenesis of Plasmodiophora brassicae in pakchoi and biological characteristic of pathogen(小白菜根肿病发生规律、发病条件及病菌生物学特性研究)[D].Chengdu:Sichuan Agricultural University(成都:四川农业大学),2009.
[4]MORGUCHI-K,KIMIZUKA-TAKAGI-C, ISHII-K,et al.A genetic map based on RAPD,RFLP isozyme,morphological markers and QTL analysis for clubroot resistance in Brassica oleracea[J]. Breeding Science,1999,49(4):257-265.
[5]WANG FZ(王芳展),LIU YP(刘亚培), ZHANG M(张梅),et al.Development of physiological,biochemical characteristics and resistant genetics during clubroot disease in crucifer crops(十字花科作物根肿病的侵染生理与抗性遗传研究进展)[J].Chinese Journal of Oil Crop Sciences(中国油料作物学报),2012,34 (2):215-224.
[6]SI J(司军),LI CQ(李成琼),XIAO CG(肖崇刚),et al.Inheritance of resistance to clubroot disease in cabbage(甘蓝根肿病抗性遗传规律的研究)[J].Acta Horticulturae Sinica(园艺学报),2003,30(6): 658-662.
[7]AYERS GW,LELACHEUR KE.Genetics of resistance in rutabaga to races of Plasmodiophora brassicae[J].Canadian Journal of Plant Science,1972,52: 897-900.
[8]DIXON GR,ROBINSON DL.The susceptibility of Brassica oleracea cultivars to Plasmodiophora brassicae(clubroot) [J].Plant Pathology,1986,35(1):101-107.
[9]RAHMAN H,SHAKIR A,JAKIR HASAN M.Breeding for clubroot resistant spring canola(Brassica napus L.)for the Canadian prairies:Can the European winter canola cv.Mendel be used as a source of resistance[J].Canadian Journal of Plant Science,2011,91(3):447-458.
[10]LAURENS F,THOMAS G.Inheritance of resistance to clubroot(Plasmodiophora brassicae wor.)in kale(Brassica oleracea ssp.acephala L.)[J].Hereditas,1993,119(3):253-262.
[11]VOORRIPS RE,VISSER DL.Examination of resistance to clubroot in accessions of Brassica oleracea using a glasshouse seedling test[J].Netherlands Journal of Plant Pathology, 1993,99(5-6):269-276.
[12]TOXOPEUS H,JANSSEN AMP.Clubroot resistance in turnip.II.The slurry screening method and clubroot races in Netherlands[J].Euphytica,1975,24 (3):751-755.
[13]TJALLINGII F.Testing clubroot resistance of turnips in netherlands and physiologic specialization of Plasmodiophora brassicae[J].Euphytica, 1965,14(1):1-22.
[14]YOSHIKAWA H.Studies on breeding of clubroot resistance in cole crops[J]. Bull Natl Res Inst Veg Ornam Plants Tea Jpn.1993,Ser A 7:1-165.
[15]DIEDERICHSEN E,ECKMANN J, SCHONDELMEIER J,et al.Genetics of clubroot resistance in Brassica napus Mendel[J].Acta Hort,2006,706: 307-311.
[16]WANG FZ(王芳展),LIU YP(刘亚培), ZHANG M(张梅),et al.Development of physiological,biochemical characteristics and resistant genetics during clubroot disease in crucifer crops(十字花科作物根肿病的侵染生理与抗性遗传研究进展)[J].Chinese Journal of Oil Crop Sciences(中国油料作物学报), 2012,34(2):215-224.
[17]MATSUMOTO E,YASUI C,OHI M,et al.Linkage analysis of RFLP markers for clubroot resistance and pigmentation in Chinese cabbage(Brassica rapa ssp.pekinensis)[J].Euphytica,1998, 104(2):79-86.
[18]SUWABE K,TSUKAZAKI H,IKETANI H,et al.Identification of two loci for resistance to clubroot(Plasmodiophora brassicae Woronin)in Brassica rapa L. [J].Theoretical and Applied Genetics, 2003,107(6):997-1002.
[19]KUGINUKI Y,AJISAKA H,YUI M,et al.RAPD markers linked to a clubroot resistance locus in Brassica rapa L.[J]. Euphytica,1997,98(3):149-154.
Responsible editor:Qingqing YIN
Responsible proofreader:Xiaoyan WU
不结球白菜抗根肿病遗传规律和分子标记
宋波,索欢,陈龙正,徐海,樊小雪,张慧,袁希汉*
(江苏省农业科学院蔬菜研究所,江苏南京210014)
[目的]探讨不结球白菜根肿病抗性的遗传规律。[方法]该研究利用引进的结球白菜与不结球白菜进行杂交,以双亲、F1、F2和BC14个世代联合群体为试材,采用苗期接种鉴定法进行抗性鉴定,并进行ⅠSSR分子标记筛选。[结果]不结球白菜根肿病抗性受1对显性基因控制。利用ⅠSSR分子标记结合BSA法获得了与不结球白菜根肿病抗性连锁的分子标记,命名为CR-873。该标记与抗性基因间的连锁遗传距离为9.72 cM。[结论]研究结果可为不结球白菜分子标记辅助育种奠定基础。
不结球白菜;抗根肿病;分子标记
江苏省自然科学基金(BK20130715);“十二五”农村领域国家科技计划子课题(2013BAD01B04-11)。
宋波(1981-),男,四川乐山人,硕士,研究方向:十字花科蔬菜遗传育种,E-mail:anybody119@sina.com。*通讯作者,研究员,硕士生导师,从事十字花科蔬菜遗传育种方面研究,E-mail:xhyuan258@163.com。
2015-03-01
修回日期 2015-05-20
Supported by Natural Science Foundation of Jiangsu Province(BK20130715);National Science and Technology Program for Rural Development during the 12thFive-Year Plan Period(2013BAD01B04-11).
*Corresponding author.E-mail:xhyuan258@163.com
Received:March 1,2015 Accepted:May 20,2015
猜你喜欢
杂志排行
Agricultural Science & Technology的其它文章
- Highly-efficient Stereo-cultivation Model in Kiwifruit Orchards Interplanting Konjak
- The Breeding of New Indica-japonica Intersubspecific Hybrid Rice Combination Chunyou 84
- Comparative Analysis between Triple Cross and Double Cross among Three Parents of Crop
- Optimization of Field Arrangement of Doublecropping Glutinous Sorghum and Soybean Intercropping Pattern
- Principal Component Analysis on Traits Related to Lodging Resistance of Plateau Japonica Rice
- Inheritance of Aroma of Good-quality Indica Type Rice PTGMS Line GHS and Its Application in Hybrid Rice Breeding
