Cloning and sequence analysis of nine novel MYB genes in Taxodiaceae plants
2014-09-06YongquanLUQingJIAZaikangTONG
Yong-quan LU · Qing JIA · Zai-kang TONG
ORIGINAL PAPER
Cloning and sequence analysis of nine novel MYB genes in Taxodiaceae plants
Yong-quan LU · Qing JIA · Zai-kang TONG
Received: 2013-12-20; Accepted: 2014-02-20
© Northeast Forestry University and Springer-Verlag Berlin Heidelberg 2014
Myeloblastosis (MYB) is one of the largest transcribed factor families in plants. To gain an overall picture of the evolution of MYB genes in relict plants, we cloned nine novel MYB genes in Taxodiaceae plants (Taxodium distichum, Taxodium ascendens, Cryptomeria japonica var. Sinensis, Cryptomeria japonica cv. Araucarioides, Cryptomer Japonica, Metasequoia glyptostroboides, Cunninghamia lanceolata, Taiwania cryptomerioides and Glyptostrobus pensilis). The deduced amino acid sequences for MYBs showed that the nine MYB proteins contained two DNA binding domains. The first domain is from amino acid position 29 to 78, wherein three tryptophanes at 33, 53 and 73 were separated by 19 amino acids, respectively. The second domain is from amino acid position 82 to 127, wherein three tryptophanes at 86, 105 and 124 were separated by 18 amino acids, respectively, whereas the first tryptophane at amino acid position 86 is replaced by a phenylalanine. The characterization of these conserved domains at nine MYBs indicated that they all belong to the R2R3-MYB group. The secondary structure analysis showed that α-helix and β-turn are the major motifs of the predicted secondary structure of MYBs. The three dimensional model of each MYB protein showed that the structure is like clip, making it more flexible and mobile. The similarities between the nine MYB proteins in Taxodiaceae were calculated. The highest identical value of 99% is between CjsMYB, CjMYB and CjaMYB, whereas the lowest value of 82% is between TaMYB and ClMYB. According to the phylogenetic tree, the distances between different genera were relatively large whereas those within genera were relatively small. As expected, accessions of the same genus formed a subgroup before being grouped with other genera.
Myeloblastosis, Taxodiaceae, clone, analysis, R2R3-MYB group
Introduction
MYB (v-myb avian myeloblastosis viral oncogene homolog), one of the largest transcribed factor families in plants, participate in plant growth development and metabolic regulation (Moyano et al. 1996). Specifically, MYB contributes to a cell’s secondary metabolic processes by taking part in regulating the transcribed activity of phenylalanine ammonia-lyase gene (PAL). MYB can regulate cell morphology by activating particular materials synthesis in epidermal cells (Wang et al. 2003). They participate in stress response by inducing the transcription of genes in the ABA pathway (Abe et al. 1997). In forestry plants, the MYB genes help regulate the formation of the tiny tube and xylem by controlling the expressions of the cinnamoyl-CoA reductase gene and cinnamyl alcohol dehydrogenase gene (Goicoechea et al. 2005). In addition, they control the formation of a cork layer by regulating the synthesis of lignin (Lopes et al. 2001; Patalaff et al. 2003).
Plant MYB proteins were classified into three major types: R2R3-MYB, with two adjacent MYB repeats domain; R1R2R3-MYB, with three adjacent MYB repeats domain; and a heterogeneous group collectively referred to as the MYB-related proteins, which usually but not always contain a single MYB domain (Rosinski and Atchley 1998; Jin and Martin 1999; Stracke et al. 2001). MYB transcription factors usually contain an incompletion repeat area of 51−52 amino acids, which include several highly conserved amino acids. The first feature within this conserved domain is several tryptophanes separated by approximately 17−19 amino acids. This characteristic plays an important role during formation of hydrophobic groups in the protein spatial structure. Sometimes, the tryptophane in this position would be replaced by other aromatic amino acids or a hydrophobic amino acid. Especially in the R2R3-MYB in plants, which contain two MYB domains, the first tryptophane in the second domain is usually replaced by leucine, isoleucine, orphenylalanine. The second feature in this conserved domain is that some conserved amino acids exist flanking each tryptophane. For example, a cluster of acidic amino acids exist in the C terminal of the first tryptophane. Those conserved amino acids are required for the formation of a helix-helix-turn-helix structure, which is functionally important for the MYB transcription factors (Bilaud et al. 1996; Braun et al. 1999; Yu et al. 2000).
In the past decade, the MYB genes have been extensively studied (Chen et al. 2006; Zhang et al. 2011; Du et al. 2012). The fact that these genes exist widely in eukaryotes suggests that they may be very ancient genes. However, only a few members of this family have been discovered in gymnosperms (Bedon et al. 2007). Despite recent large-scale gene discoveries for conifer trees such as pine and spruce (Kirst et al. 2003; Pavy et al. 2005), only a few regulatory gene families have been systematically studied in the gymnosperm class. One example is the Taxodiaceae family, which are relict plants from Cretaceous period. The family was an important component in forest vegetation of the northern hemisphere from the late Cretaceous to the mid-tertiary eras approximately 115 to 30 million years ago. In the Tertiary and Pleistocene, however, the family underwent a widespread reduction resulting in the present-day relictual genera with restricted distributions (Sehlabraum and Tsuehiay 1984). At present, the Taxodiaceae consists of 10 arborescent genera: seven of which are monotypic, and the remainder are olictypic genera consisting of only two or three species, resulting in a wide range of morphological characteristics (Wu 1998). Up to now, there are few MYB gene was cloned and the characteristics of MYB genes within Taxodiaceae family is still unambiguous. As an important transcript in stress response, when the Taxodiaceae family went through the great changes of climate in the Tertiary and Pleistocene, MYB should play a role to cope with weather changes. Therefore, to study the evolution of MYB genes within this family would be necessary.
Materials and methods
Plant materials
Fresh leaves of nine Taxodiaceae accessions representing six genera were collected from Hangzhou Plant Garden, Zhejiang, China (Table 1). The natural distribution of these six genera are extremely scattered, forming interrupted or isolated distribution.
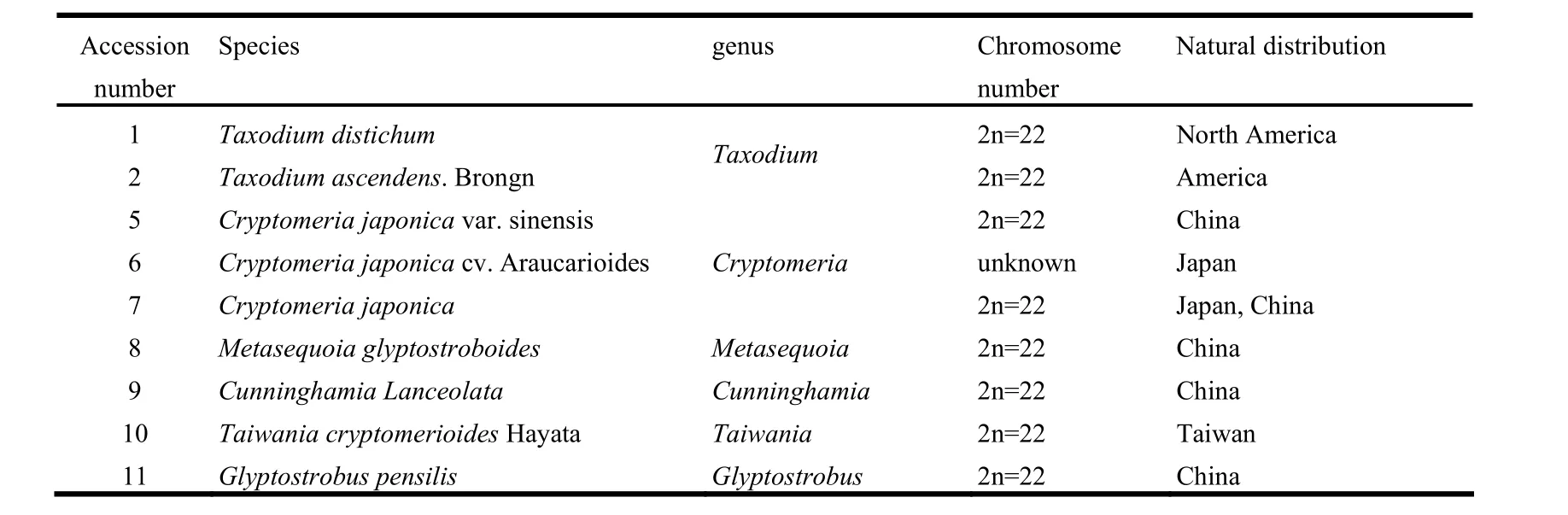
Table 1: Accessions used for Amplified Consensus Genetic Marker analysis
MYB genes clone
RNAs were isolated from 500 mg of leaves from each accession, using the CTAB method with modification (Murray and Thompson 1980). They then were reversed into cDNA (coding DNA) as templates for PCR reactions. Primers of amplified consensus genetic markers targeted at the MYB gene (Lu et al. 2013) were used in the first PCR reaction.
PCR was performed in 20 μL reactions containing 50 mg of template DNA; 0.5 mol/L of each target primer (see Table 2); 200 mol/L of each dNTP; 1.5 mmol/L of MgCl2; 1 unit of Pfu DNA polymerase; and 2 L of 10×PCR reaction buffer. A touchdown PCR program (Don et al. 1991) was used: five min at 95°C; 10 cycles of: 30 s at 95°C, 30 s at 58°C minus 0.3°C per cycle, one min. at 72°C; 20 cycles of: 30 s at 95°C, 30 s at 55°C, one min at 72°C; and seven min at 72°C for a final extension. The PCR products were cloned into a pGEMT-vector, and the positive clones were picked out. The T-vector then was identified by restriction endonuclease digestion.
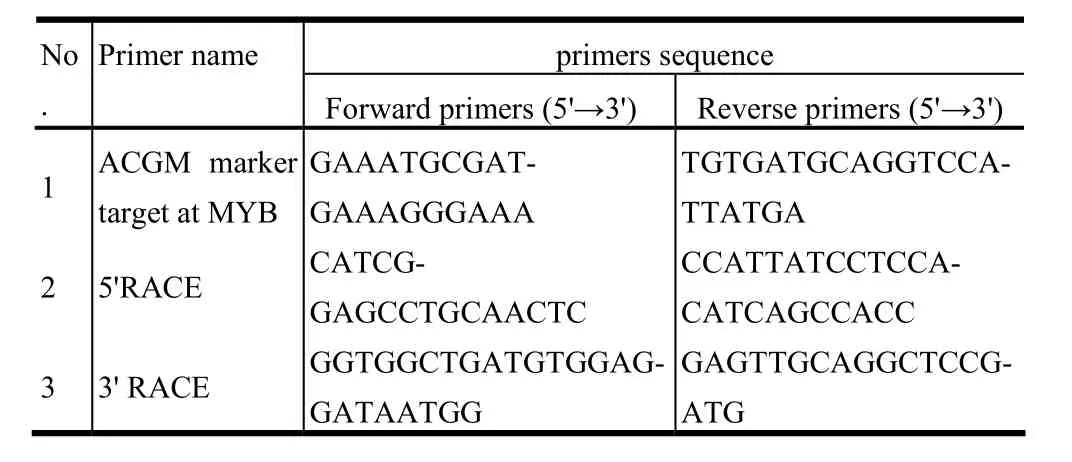
Table 2: Primers for target genes
The correct clones were selected for sequencing. According to the sequencing results, 5' and 3' RACE (rapid amplification of cDNA ends) primers were designed (see Table 3). Following the RACE manual, the 5' and 3' fragments of MYB genes were obtained, respectively.
We then designed primers to clone coding sequence (CDS) of MYB genes from nine accessions, respectively. The primers for gene cloning are in Table 3. All primers used were synthesized by the Nanjing Jinsirui Biological Engineering & Technology Company (Nanjing, China). Electrophoreses of PCR products were run on 1.0% agarose gel. The target products were purified and cloned into pGEM-T vector (Promega, USA) for sequencing (Sangon, Shanghai, China).
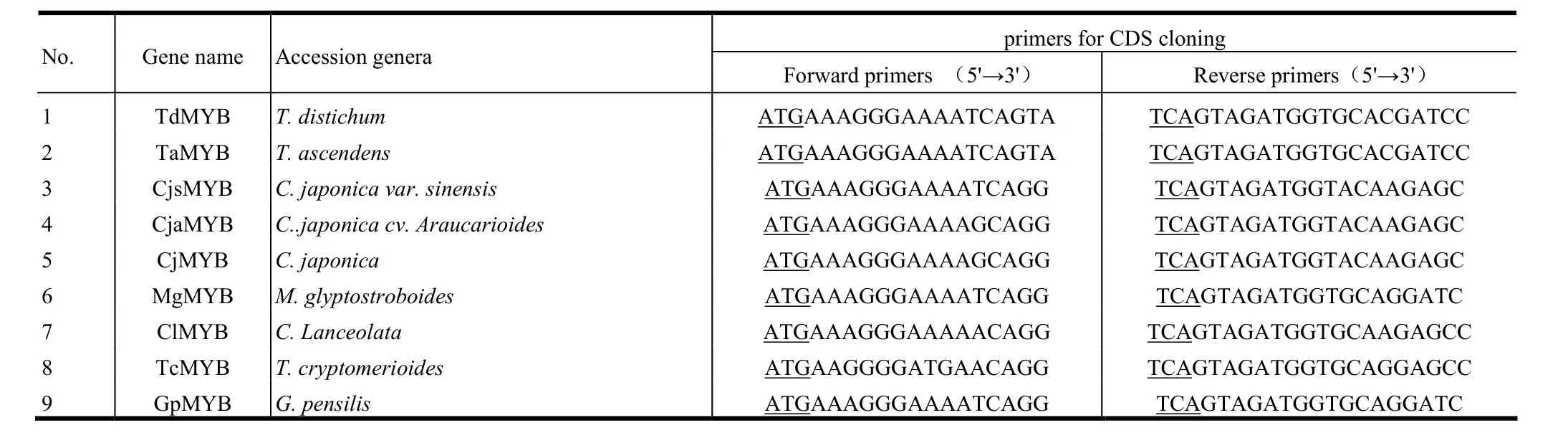
Table 3: Primers for cloning genes
Characterization of the deduced MYB protein
Amino acid sequences of MYBs were deduced using DNAman software. The isoelctric points and the molecular weights of the deduced MYB polypeptides were calculated with the ProtParam tool (http://web.expasy.org/protparam). The conserved domains of MYBs were also predicted by SMART (http://smart.embl-heidelberg.de). Secondary structure of each MYB protein was checked in GOR (http://npsa-pbil.ibcp.fr). Finally, the three-dimensional model of each MYB was set up with 3D-pssm of ExPASy (http://en.wikipedia.org).
Homologous analysis
The nine MYBs from Taxodiaceae plants obtained in this study, as well as with MYBs from other plant species, including Hordeum vulgare, Triticum aestivum, Solanum lycopersicum, Lolium temulentum, Medicago truncatula and Arabidopsis thaliana, were alignment by Clustal X. Phylogenetic tree was constructed using Mega 5.
Results
Analysis of MYB sequence
Among the sequences, the start codon and termination codon were all obtained in those nine MYB nucleotides, respectively. Database search with BlastN at NCBI (www.ncbi.nlm.nih.gov) showed that the nine MYB nucleotide sequences had a high similarity (> 70%) with other MYB genes from both gymnosperm and angiosperm plant species. Those results showed that the full coding sequences of nine MYB genes in Taxodiaceae plants were obtained, respectively (Fig. 1). The lengths of the nine MYB genes range from 1371−1392 bp (Fig. 2). The lengths of deduced amino acids range from 456 to 463 (Fig. 3). Further analysis showed that the deduced amino acid sequences of MYBs contained several variation sites. According to sequences of multiple alignments with Clustal X, serine deletion mutations were found at amino acid position 21 in MgMYB, GpMYB, ClMYB, and TcMYB, and continuous amino acid deletion at 256 in TcMYB. Serine insertion mutations were found at amino acid position 225 in TdMYB and TaMYB, at 190 in ClMYB, at 341 in ClMYB and TcMYB, respectively.
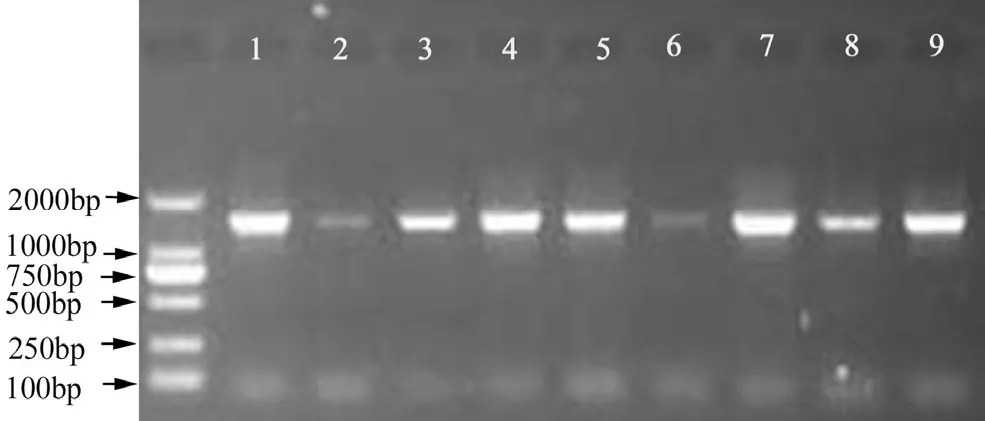
Fig. 1: PCR resules of 9 MYB genes
Conserved domain analysis of the MYB protein
MYB transcription factors usually contain an incompletion repeat area of 51−52 amino acids, which include several highly conservative amino acids. In our study, the deduced amino acid sequences for MYBs showed that each MYB protein contained two DNA binding domains (Chen and Wang 2002). The first domain is from amino acid position 29 to 78, wherein three tryptophanes (W) at 33, 53 and 73 were separated by 19 amino acids, respectively (Fig. 3). The second domain is from amino acid position 82 to 127, wherein three tryptophanes (W) at 86, 105 and 124 were separated by 18 amino acids, respectively, whereas the first tryptophane at position 86 is replaced by a phenylalanin (F). The characterization of these conserved domains at nine MYB indicates that they all belong to R2R3-MYB group.
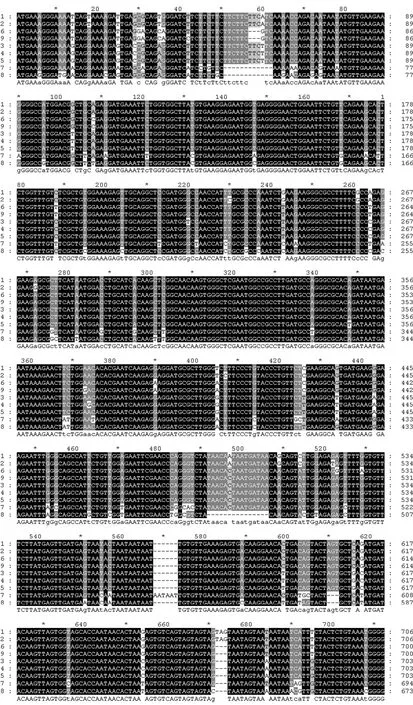
Fig. 2: Multiple alignment of 9 MYB genes

Continued Fig. 2: Multiple alignment of 9 MYB genes
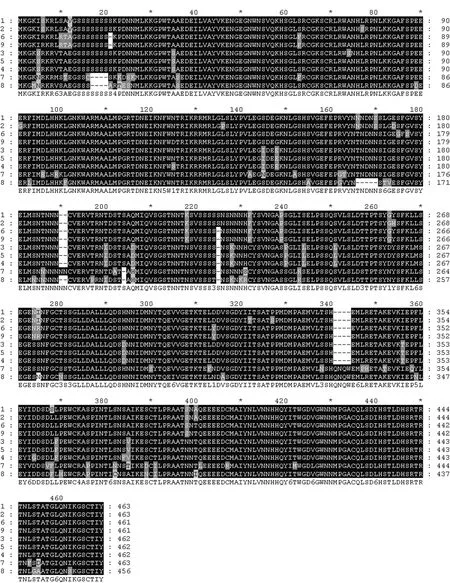
Fig. 3: Multiple alignment of MYB protein. Note: Conserved domains are underlined.
Prediction of MYB protein
The secondary structures analysis showed that α-helix and β-turn are the major motifs of predicted secondary structure of MYBs (Table 4). The numbers of α-helix and β-turn among nine MYBs range from 7 to 10 and 19 to 25, respectively. The variations of secondary structure between the nine MYBs are resulted from the amino acid sequences variations. Mutation of serine (S) to phenylalanine (F) at amino acid position 64 in MgMYB, ClMYB,and GpMYB result in a β-turn insertion in these positions, respectively. Whereas, mutation of leucine (L) to serine (S) at position 78 in TaMYB, as well as alanine (A) to serine (S) at position 75 in CjsMYB, resulted in a α-helix deletion in these positions, respectively. Although some variations exist among the nine MYB proteins, their secondary structures are similar, which showed the conservation of MYBs in evolution. The three-dimensional model of each MYB protein was set up with 3D-pssm of ExPASy (Fig. 4). The two DNA binding domains of MYBs can be seen clearly. The structure of each MYB is like a clip, resulting in the MYB transcription factors having more flexibility and mobility when binding target DNA.
Homologous analysis
Database search with BlastP in NCBI and multi-alignment Clustal X 1.83 showed that the deduced MYB proteins had similarity (from 74% to 78%) with MYBs from other plant species (Fig. 5). The similarities among MYB proteins we obtained in Taxodiaceae were calculated. The highest identical value of 99% is between CjsMYB, CjMYB and CjaMYB, whereas the lowest value of 82% is between TaMYB and ClMYB. The hylogenetic tree was constructed with Mega5 (Fig. 6). According to the tree, the distances between different genera were relatively large whereas those within genera were relatively small. As expected, accessions of the same genus formed a subgroup before being grouped with other genera.
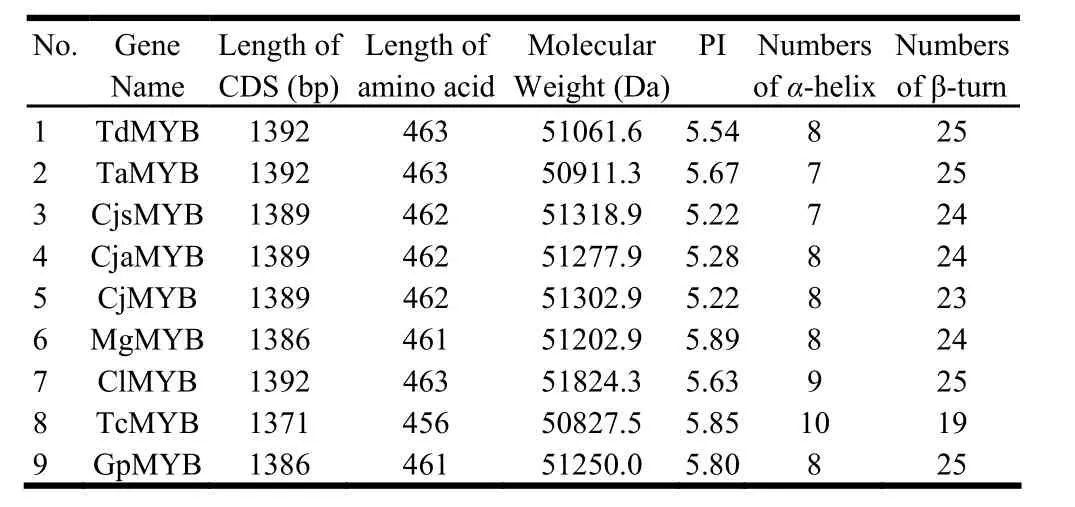
Table 4: Results of sequence analysis
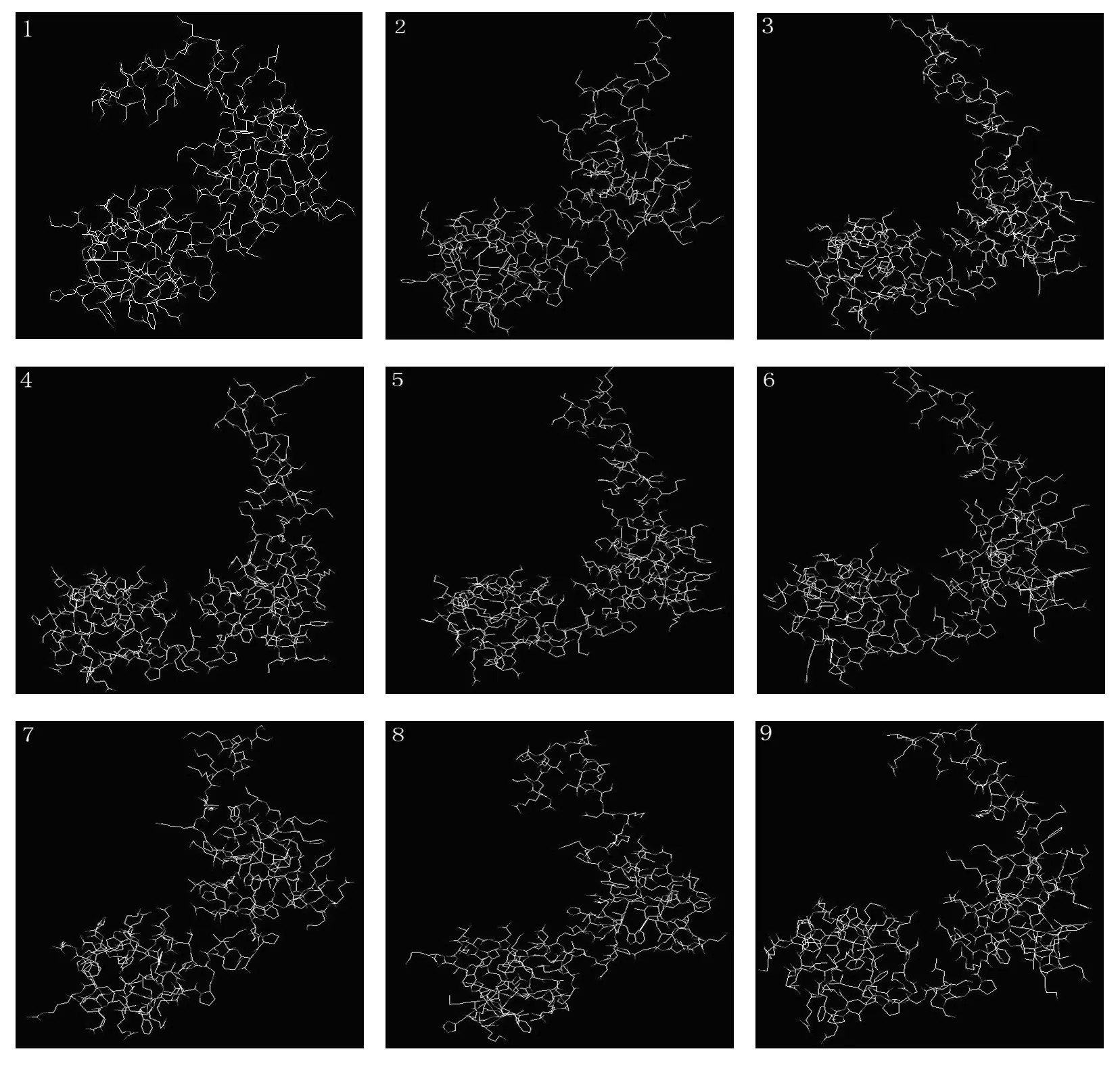
Fig. 4: Predict the three-dimensional of MYB protein. Note: The number one to nine correspond to the accession number in Table 1.

Fig. 5: Multiple alignment of MYB protein among different species: Hordeum vulgare (X87690.1), Triticum aestivum (AY615200.1), Solanum lycopersicum (NM_001247433.1), Lolium temulentum (AF114162.1), Medicago truncatula (XM_003589214.1), Arabidopsis thaliana (NM_118827.1); Numbers correspond to accessions in Table 1.
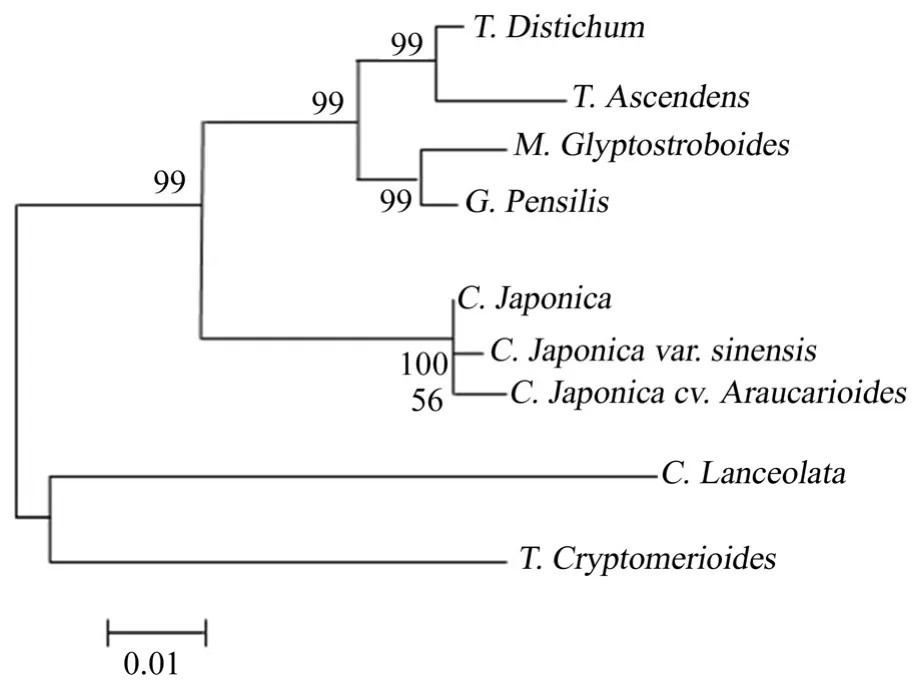
Fig. 6: Phylogenetic tree of MYB protein
Discussion
To achieve an overall picture of this gene super-family in relict plants, we have gained nine accessions of R2R3-MYB genes in the Taxodiaceae family. In the past decade, the R2R3-MYB genes have been extensively studied. They were reported to be involved in many physiological and biochemical processes. The members in this group are numerous. Chen et al. (2006) identified 198 genes in the MYB superfamily from an analysis of the complete Arabidopsis genome sequence; among them, 126 (63.6%) are R2R3-MYB genes. However, bio-information on Taxodiaceae plants is limited and the genome of these plants remains unknown. We designed primers according to a reserved sequence of MYB domain and obtained MYB genes from nine accessions of the Taxodiaceae family. The nine MYB genes all belong to R2R3-MYB, which indicated that R2R3-MYB is the main type in these plants. The R2R3-MYB group of transcription factors is one of the largest regulatory gene families known in plants (Cedroni et al. 2003). The MYB domain is well conserved among plants, yeast and animals (Lipsick 1996). Therefore the origin and evolutionary history of members of this large gene family are of interest. Similarities in the nine MYB proteins obtained in Taxodiaceae also closely corresponded with MYBs from other plant species, implicating the evolutionary conservation of MYB gene. The conserved domains of R2R3-MYB in Taxodiaceae plants are exactly the same as the domains that are necessary for MYB functions. According to multiple sequences alignment of the nine MYB genes, the amino acids sequences of T. cryptomerioides and C. lanceolata showed more differences, mainly with insertion and deletion mutations. For example there are four continuous (SSSS) inserts at the amino acid position 17−20, and four amino acids (QNQW) insert mutations at positions 341−344. These variations made T. cryptomerioides and C. lanceolata cluster into one group and showed that the two species have very similar genetic relationship.
Abe H, Yamaguchi-Ssinozake K, Urao T, lwasaki T, Hosokawa D, Shinozaki K. 1997. Role of Arabidopsis MYC and MYB homologs in drought and abscisic acid-regulated gene expression. Plant Cell, 9: 1859−1868.
Bedon F, Grima-Pettenati J, John M. 2007. Conifer R2R3-MYB transcription factors: sequence analyses and gene expression in wood-forming tissues of white spruce (Picea glauca). BMC Plant Biology, 7:17−33
Bilaud T, Koering CE, Binet-Brasselet E, Ancelin K, Pollice A, Gasser SM, Gilson E. 1996. The telobox, a myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res, 24: 1294–1303.
Braun EL, Grotewold E. 1999. Newly discovered plant c-myb2like genes rewrite the evolution of the plant myb gene family. Plant Physiol, 121: 21–24.
Cedroni ML, Cronn RC, Adams KL, Wilkins TA, Wende JF. 2003. Evolution and expression of MYB genes in diploid and polyploid cotton. Plant Molecular Biology, 51: 313–325.
Chen J, Wang ZY. 2002. Progress in the study of plant MYB transcription factors. Journal of Physiology and Molecular Biology, 28(2): 810–88. (in Chinese with English abstract)
Chen YH, Yang XY, He K, Liu MH, Li JG, Gao ZF, Lin ZQ, Zhang YF, Wang XX, Qiu XM, Shen YP, Zhang L, Deng XH, Luo JC, Deng XW, Chen ZL, Gu HY, Qu LJ. 2006. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Molecular Biology, 60: 107–124.
Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. 1991. ‘Touchdown’PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res, 19: 4008–4008.
Du H, Yang SS, Liang Z, Feng BR, Liu L, Huang YB, Tang YX. 2012. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biology, 12: 106–127.
Goicoechea M, Lacombe E, Legay S. 2005. EgMYB2-a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant Journal, 43(4): 553–567.
Jin H, Martin C. 1999. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol, 41: 577–585.
Kirst M, Johnson AF, Baucom C, Ulrich E, Hubbard K, Staggs R, Paule C, Retzel E, Whetten R, Sederoff R. 2003. Apparent homology of expressed genes from wood-forming tissues of loblolly pine (Pinus taeda L.) with Arabidopsis thaliana. Proc Natl Acad Sci USA, 100(12): 7383–7388.
Lipsick JS. 1996. One billion years of Myb. Oncogene, 13: 223–235.
Lopes M, Barros A, Neto C. 2001. Variability of cork from Portuguese Quercus suber studied by solidstate C-13-NMR and FTIR spectroscopies. Biopolymers, 62(5): 268–277.
Lu YQ, Jia Q, Tong ZK. 2013. Amplified consensus genetic markers in Taxodiaceae based on Cryptomeria japonica ESTs data. Journal of Forestry Research, 24(3): 503–508.
Moyano E, Martin EZ, Garcia JF, Martin C. 1996. Apparent redundancy in myb-gene function provides gearing for the control of flavonoid bio-synthesis in Antirrhinum flowers. Plant Cell, 8: 1519–1532.
Murray MG, Thompson WF. 1980. Rapid isolation of high molecular-weight plant DNA. Nucleic Acids Res, 8: 4321–4325
Patalaff A, Mcinnis S, Courtenay A. 2003. Characterisation of a pine MYBthat regulates lignification. Plant Journal, 36(6): 743–754.
Pavy N, Laroche J, Bousquet J, Mackay J. 2005. Large-scale statistical analysis of secondary xylem ESTs in pine. Plant Mol Biol, 57(2): 203–224.
Rosinski JA, Atchley WR. 1998. Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. J Mol Evol, 46: 74–83.
Sehlabraum SE, Tsuehiay T. 1984. Cytotaxonomy and phylogeny in certain species of Taxodiaceae. Syst Evol, 147: 29–54.
Stracke R, Werber M, Weisshaar B. 2001. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol, 4: 447–456.
Wang XQ, Chen BJ, Yin LP. 2003. The plant MYB transcription factors. Biotechnology Information, 2: 22–25.
Wu ZY. 1998. Flora of China, Volume 7: Taxodiaceae. Beijing: China Science Press,
Yu EY, Kim SE, Kim JH, Ko JH, Cho MH, Chung IK. 2000. Sequence-specific DNA recognition by the Myb-like domain of plant telomeric protein RTBP1. J Biol Chem, 275: 24208–24214.
Zhang F, Liu X, Zuo KJ, Sun XF, Tang KX. 2011. Molecular cloning and expression analysis of a novel SANT/MYB gene from Gossypium barbadense. Mol Biol Rep, 38: 2329–2336.
DOI 10.1007/s11676-014-0527-1
Project funding: This work was funded by the Natural Science Foundation of China (30800879) and project 2009R50035 supported by Forest Seedling Industry Innovative Team of Zhejiang province in China.
The online version is available at http://www.springerlink.com
The Nurturing Station for the State Key Laboratory of Subtropical Silviculture, Zhejiang Agriculture and Forestry University, Lin’an 311300, China. Email: luyongquan@126.com
Corresponding editor: Hu Yanbo
杂志排行
Journal of Forestry Research的其它文章
- Growth and yield of two grain crops on sites former covered with eucalypt plantations in Koga Watershed, northwestern Ethiopia
- Full length cDNA cloning and expression analysis of annexinA2 gene from deer antler tissue
- Bamboo resources of Sikkim Himalaya: diversity, distribution and utilization
- Implications of crude oil pollution on natural regeneration of plant species in an oil-producing community in the Niger Delta Region of Nigeria
- Enhancement of seed germination in Macaranga peltata for use in tropical forest restoration
- Bio-amelioration of alkali soils through agroforestry systems in central Indo-Gangetic plains of India
