The Appearance of Ulva laetevirens (Ulvophyceae, Chlorophyta) in the Northeast Coast of the United States of America
2014-04-20MAOYunxiangJangKyunKimRoderickWilsonandCharlesYarish
MAO Yunxiang, Jang Kyun Kim, Roderick Wilson and Charles Yarish
1) Department of Ecology and Evolutionary Biology, University of Connecticut at Stamford, Stamford CT 06901-2315, USA
2) Key Laboratory of Marine Genetics and Breeding (MOE), College of Marine Life Sciences, Ocean University of China, Qingdao 266003, P. R. China
3) Department of Marine Sciences, University of Connecticut at Stamford, Stamford CT 06901-2315, USA
The Appearance of Ulva laetevirens (Ulvophyceae, Chlorophyta) in the Northeast Coast of the United States of America
MAO Yunxiang1),2),*, Jang Kyun Kim3), Roderick Wilson1), and Charles Yarish1)
1) Department of Ecology and Evolutionary Biology, University of Connecticut at Stamford, Stamford CT 06901-2315, USA
2) Key Laboratory of Marine Genetics and Breeding (MOE), College of Marine Life Sciences, Ocean University of China, Qingdao 266003, P. R. China
3) Department of Marine Sciences, University of Connecticut at Stamford, Stamford CT 06901-2315, USA
Introduced species may outcompete or hybridize with native species, resulting in the loss of native biodiversity or even alteration of ecosystem processes. In this study, we reported an alien distromatic Ulva species, which was found in an embayment (Holly Pond) connected with Long Island Sound, USA. The morphological and anatomical observations in combination with molecular data were used for its identification to species. Anatomy of collected specimens showed that the cell shape in rhizoidal and basal regions was round and the marginal teeth along the basal and median region were not found. These characteristics were primarily identical to the diagnostic characteristics of Ulva laetevirens Areschoug (Chlorophyta). The plastid-encoding tufA and nucleus-encoding ITS1 were used for its molecular identification. Phylogenetic analysis for the tufA gene placed the specimens from Holly Pond in a well-supported clade along with published sequences of U. laetevirens identified early without any sequence divergence. In ITS tree, the sample also formed well-supported clades with the sequences of U. laetevirens with an estimated sequence divergence among the taxa in these clades as low as 1%. These findings confirmed the morpho-anatomical conclusion. Native to Australia, this species was reported in several countries along the Mediterranean coast after the late of 1990s. This is the first time that U. laetevirens is found in the northeast coast of United States and the second record for Atlantic North America.
Ulva laetevirens Areschoug; introduced seaweed; Long Island Sound, USA; ITS; tufA
1 Introduction
Introduced seaweeds are a growing and imminent threat throughout the world’s oceans, altering ecological structure and causing economic harm (Nyberg and Wallentinus, 2005; Mathieson et al., 2008). Based upon floristic and molecular investigations, 23 introduced seaweeds are presently known from the Northwest Atlantic, including 3 green, 4 brown and 16 red algae (Mathieson et al., 2008; Thornber et al., 2009; Hofmann et al., 2010; Schneider, 2010). Many Ulva species are notorious biofoulers of ships’ hulls and ballast waters, making them among the most commonly transported and widely introduced species of macroalgae (Nelson et al., 2007). Ulva laetevirens Areschoug, a green seaweed native to south Australia (Areschoug, 1854), was recently reported in several Mediterranean coastal countries including Italy (Furnari et al., 1999; Rindi et al., 2002; Sfiso, 2010), Slovenia (Rindi and Battelli, 2005), Israel (Einav, 2007), Greece and Cyprus (Christia et al., 2011). It was also recently reported in the Atlantic coast of Canada (Kirkendale et al., 2013). Here we reported the appearance of Ulva laetevirens Areschoug in the Long Island Sound, northwest Atlantic Ocean of USA.
2 Materials and Methods
2.1 Location and Habitats
The samples were collected from two sites of Holly Pond (Stamford, Connecticut, 41˚2´57.87´´N, 73˚29´ 55.66´´W) on Jun 21, 2011. In general, temperature and salinity of site 1 are more variable than those of site 2. In site 1, plants are often exposed to air for 9–10 h during the tidal cycle. This site is rapidly influenced by fresh water runoff, with the salinity ranging from 0.0 to 26.0 (mean= 18.3). Water temperature ranges from -1.0℃ in January to 29.0℃ in August. The substratum of site 1 is dominated by mud, and Ulva plants attach to small pebbles or oyster shells. Site 2 is situated just above the dam. Salinity ranges from 2.0 to 28.0 (mean = 23.4 ± 5.48) and temperature ranges from -2.0℃ to 26.5℃. The bottom is characterized by shell fragments, stones and fine sediment. Currents are minimal at both sites (Mariani, 1983).
2.2 DNA Extraction and PCR
Following the collection, each specimen was cleanedand pressed on a herbarium paper as a voucher, while a subsample was frozen for DNA extraction.
The freshly-frozen portion was ground in liquid nitrogen. DNA was extracted using a Qiagen DNeasy Plant Mini Kit (Valencia, CA, USA) following the manufacturer’s protocol. The PCR mixture was prepared by mixing 25 μL of AmpliTaq Gold PCR Master Mix (Applied Biosystems, Inc., Carlsbad, CA, USA) containing AmpliTaq Gold with 0.5 μL of forward primer (10 μmol L-1), 0.5 μL of reverse primer (10 μmol L-1), 1 μL of genomic DNA as template (5–10 ng) and 23 μL of distilled water for a total volume of 50 μL. PCR amplifi cation was carried out in a GeneAmp PCR System 9600 (Applied Biosystems, Inc., Carlsbad, CA, USA). Primers used to amplify and sequence ITS and tufA are listed in Table 1. PCR profile for amplifying ITS was as follows: an initial denaturation at 94℃ for 5 min, followed by 1 min at 94℃ and 3 min at 60℃ for 30 cycles, and a final 10 min extension at 60℃ (Blomster et al., 1998). PCR condition for amplifying tufA included an initial 4 min denaturation at 94℃, followed by 38 cycles of amplification including denaturing at 94℃ for 1 min, annealing at 45℃ for 30 s, extending at 72℃ for 1 min, and an extra extension at 72℃ for 7 min (Saunders and Kucera, 2010).
Amplification success was evaluated using gel electrophoresis in a 1.0% agarose gel. PCR products were purif i ed using a Qiagen PCR product purif i cation kit and sequenced in both directions using an ABI 3130XL Genetic Analyzer (Carlsbad, CA, USA). The DNA sequences were trimmed using Chromas 2.22 (Technelysium Pty Ltd, Tewantin, Queensland, Australia). Sequences alignment and phylogenetic analysis were conducted using MEGA version 5.05 (Tamura et al., 2011).

Table 1 Primers used in this study for PCR amplification and sequencing
3 Results
3.1 Morphology and Anatomy
Specimens of U. laetevirens were collected from both sites (1# and 2#) of Holly Pond (Fig.1). Thalli of U. laetevirens are foliose (Fig.2), which naturally grow singly or in small clumps fixed to the hard substratum with rhizoidal. The cell shape and blade thickness vary from rhizoidal parts to apical regions. In surface view, the rhizoidal cells are difficult to clearly distinguish from the normal cells (Fig.3) in size and color, which is different from the confusable species U. rigida whose rhizoidal cells appears thicker and darker (Sfriso, 2010). The cell shape in rhizoidal and basal regions is round, whereas those in median and apical regions are polygonal or quadrangular. Marginal teeth along the basal and median regions were not found in this study (Fig.3). In cross sections, the thickness reaches 100–120 μm in the rhizoidal region and decreases to 30– 40 μm in the apical region (Fig.4). The rhizoidal and basal cells are distinctly cylindrical or conical, and their height is 2–3 times of their diameter, which is an important characteristic for determination of U. laetevirens (Sfriso, 2010). A cross-section from the rhizoidal region reveals a thick band of secondary rhizoids. In the median and apical areas, the cell length is not distinctive, but they have a conical shape as they taper towards the thallus surface.

Fig.1 Map of Long Island Sound and locations where specimens of Ulva laetevirens were found (✵).

Fig.2 Blade of U. laetevirens from Holly Pond. Scaled to a centimeter ruler.
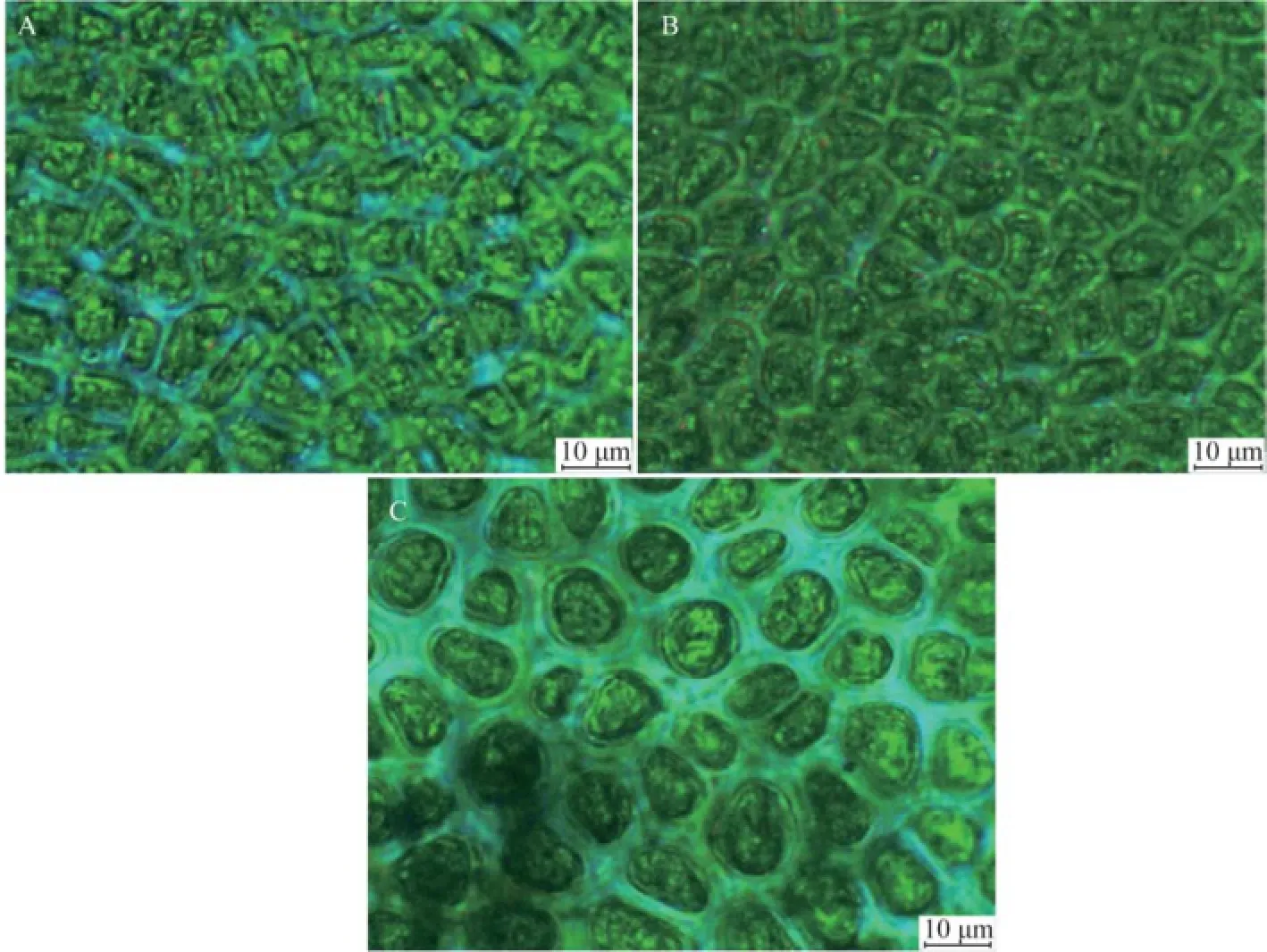
Fig.3 Microscopic surface views of U. laetevirens (scale bar =10 μm). A, Apical region; B, Median region; C, Basal region.
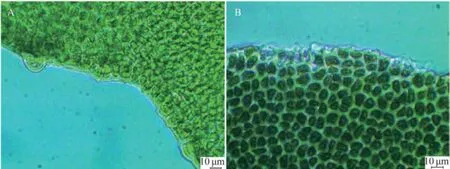
Fig.4 Marginal views of U. laetevirens (scale bar =10 μm). A, Median region; B, Basal region.
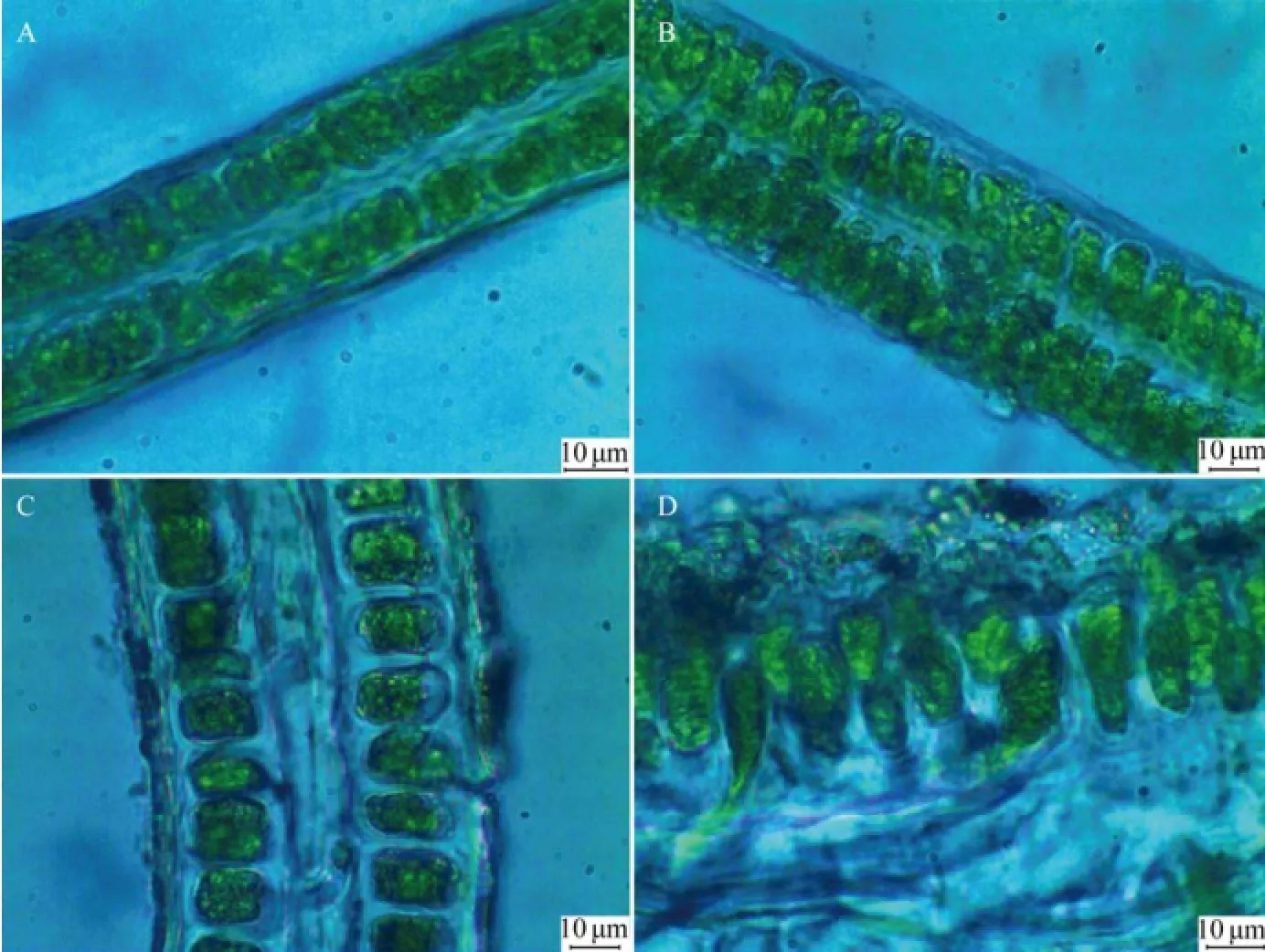
Fig.5 Microscopic cross sections of U. laetevirens (scale bar =10 μm). A, Apical region; B, Median region; C, Basal region; D, Rhizoidal region.
3.2 Molecular Evidences
We sequenced partial plastid elongation factor gene, tufA, which has recently been developed as a routine barcode of green algae (Saunders et al., 2010). After trimming the ambiguous nucleotides, the sequence length of tufA from voucher UCCY20110621P03 was 815 bp (JQ048942.1) and that was 780 bp for voucher UCCY 20110621P05 (JQ048943.1). The alignment showed that the two sequences were identical at 780 aligned sites. All available accessions of Ulva tufA genes in GenBank were collected with duplicates removed. In total, 746 sites among 35 tufA sequences were aligned, which represented 15 Ulva species, and used for reconstructing the phylogenetic tree. Molecular phylogenetic data for tufA gene placed these two specimens from Holly Pond in a well-supported clade along with published sequences of U. laetevirens (HQ610428.1) identified early without any divergence (Fig.6).
JQ048946.1 and JQ048947.1 were assigned to the ITS sequence of voucher UCCY20110621P03 and UCCY 20110621P05, respectively. The two ITS sequences are the same in length (516 bp). Alignment shows that the two ITS sequences are identical each other. There are quite a number of ITS sequences for Ulva in Genbank. The following criteria were used for picking up the representatives from GenBank for constructing ML tree: i) higher value of query cover and similarity value (>90%) against JQ048946.1; ii) with foliose thalli; iii) naturally distributing along the coast of nothern Atlantic. In ITS ML trees (Fig.7), voucher UCCY20110621P03 and UCCY20110621P05 form well-supported clades with sequences of U. laetevirens, as well as a mixture of Gen-Bank accessions given as U. amoricana, U. rigida and U. scandinavica. The estimated sequence divergence between the taxa in these clades was as low as 1%.

Fig.6 Unrooted maximum-likelihood (ML) tree for plastid-coding elongation factor gene (tufA) sequence data with ML bootstrap support values.

Fig.7 Unrooted phylogram generated with maxima-likelihood analysis from the ITS for samples included in this study.
4 Discussion
Holly Pond locates approximately 50 km east of New York Harbor. Noroton River flows into Holly Pond and empties into Long Island Sound (LIS) (Fig.1). It has been estimated that the pond refreshes only 30% of its total volume daily because of a dam constructed in 1960, which is about 80 cm higher than mean sea level (Harris, 1973). Reduced flushing of the pond and accumulation of pollutants associated with the sediments result in eutrophic waters. From the late of 1970s, Ulva species inhabiting Holly Pond have been intensively and systematically investigated. Mariani (1983) described four ‘types’ of Ulva species in Holly Pond. Type 1 appears morphologically similar to U. lactuca; type 2, type 3 and type 4 conform to the description of U curvata, U. rotundata, U. laetevirens, respectively. Ulva rigida and U. laetevirens are difficult to be accurately identified, as they display few distinctive diagnostic features, as well as large degrees of morphological plasticity within these characters. The cell features in the cross section of the rhizoidal and basal regions are considered to be a diagnostic character (Sfriso, 2010). In the rhizoidal, and sometimes in the basal region, U. rigida has large, rectangular cells. These cells are taller and narrower in other regions. In contrast, in the rhizoidal and basal regions of U. laetevirens, the cells are tall and narrow, with a conical shape (Phillips, 1988). In the specimens collected from Holly Pond, the cell shapes in the rhizoidal and basal regions are identical with the diagnostic characters of U. laetevirens described by Sfriso (2010). Marginal teeth along the basal and median region were not found in this study, which has been used as a morphological character of U. rigida (Innes et al., 1981; Hoeksema and van den Hoek, 1983) to distinguish these two species, even though it may not be adequate (Philips, 1988).

Fig.8 Distribution of Ulva laetevirens in the world drawn by referring to the documented early (Furnari et al., 1999; Rindi et al., 2002; Sfiso, 2010; Rindi and Battelli, 2005; Einav, 2007; Christia et al., 2011; Kirkendale et al., 2013).
Ulva laetevirens was originally described by J. E. Areschoug in 1854 based on the specimen collected from the Port Phillip, Victoria, Australia. From the late 1990s, this species has been reported in several Mediterranean coastal countries. Recently, this species was found in New Brunswick, Canada (Kirkendale et al., 2013). Based on the information available, we could infer the expanding tendency of this species (Fig.8). But for fully understanding its migration pattern, more specimens from North America, Mediterranean Seas and Australia should be collected to conduct the molecular genetic diversity analysis.
Acknowledgements
This work was partly funded by the China Scholarship Council. The authors wished to acknowledge supports of this work by the Connecticut Sea Grant College Program (NA10OAR4170095; CT Sea Grant R/A38), and from the Perkin Elmer research fund at the University of Connecticut, Perkin Elmer Analytical Division of E, G & G, Wellesley, MA, USA.
Areschoug, J. E., 1854. Phyceae novae et minus cognitae in maribus extraeuropaeis collectae. Nova Acta Regiae Societatis Scientiarum Upsaliensis, Ser. 3, 1: 329-372.
Blomster, J., Maggs, C. A., and Stanhope, M. J., 1998. Molecular and morphological analysis of Enteromorpha intestinalis and E. compressa (Chlorophyta) in the British Isles. Journal of Phycology, 34: 319-340.
Christia, C., Tziortzis, I., Fyttis, G., Kashta, L., and Papastergiadou, E., 2011. A survey of the benthic aquatic flora in transitional water systems of Greece and Cyprus (Mediterranean Sea). Botanica Marina, 54 (2): 169-178.
Einav, R., 2007. Seaweeds of the Eastern Mediterranean Coast. A. R. G. Gantner Verlag, Ruggell, Liechtenstein, 266pp.
Fama, P., Wysor, B., Kooistra, W., and Zuccarello, G. C., 2002. Molecular phylogeny of the genus Caulerpa (Caulerpales, Chlorophyta) inferred from chloroplast tufA gene. Journal of Phycology, 38: 1040-1050.
Furnari, G., Cormaci, M., and Serio, D., 1999. Catalogue of the benthic marine macroalgae of the Italian coast of the Adriatic Sea. Bocconea, 12: 1-214.
Guiry, M. D., and Guiry, M. G., 2012. AlgaeBase. World-wide Electronic Publication, National University of Ireland, Galway. http://www.algaebase.org.
Hayden, H. S., Blomster, J., Maggs, C. A., Silva, P. C., Stanhope, M. J., and Waaland, J. R., 2003. Linnaeus was right all along: Ulva and Enteromorpha are not distinct genera. European Journal of Phycology, 38: 277-294.
Hofmann, L. C., Nettleton, J. C., Neefus, C. D., and Mathieson, A. C., 2010. Cryptic diversity of Ulva (Ulvales, Chlorophyta) in the Great Bay Estuarine System (Atlantic USA): Introduced and indigenous distromatic species. European Journal of Phycology, 45: 230-239.
Inderjit, C. D., Ranelleti, M., and Kaushik, S., 2006. Invasive marine algae: An ecological perspective. Botanical Review, 72: 153-178.
Innes, D. J., Mariani, E. C., and Yarish, C., 1981. Use of electrophoretic markers to distinguish species of Ulva and Enteromorpha. Phycologia, 20: 107.
Hoeksema, B. W., and van den Hoek, C., 1983. The taxonomy of Ulva (Chlorophyceae) from the coastal region of Roscoff (Brittany, France). Botanica Marina, 26: 65-86.
Kirkendale, L., Saunders, G. W., and Winberg, P., 2013. A molecular survey of Ulva (Chlorophyta) in temperate Australia reveals enhanced levels of cosmopolitanism. Journal of Phycology, 49 (1): 69-81.
Loughnane, C. J., McIvor, L. M., Rindi, F., Stengel, D. B., and Guiry, M. D., 2009. Morphology, rbcL phylogeny and distribution of distromatic Ulva (Ulvophyceae, Chlorophyta) in Ireland and southern Britain. Phycologia, 47: 416-429.
Manhart, J. R., 1994. Phylogenetic analysis of green plant rbcL sequences. Molecular Phylogenetics and Evolution, 3: 114-127
Mariani, E. C., 1983. A Field and Cultural Study of Ulva in Western Long Island Sound. University of Connecticut, Storrs, 34-36.
Mathieson, A. C., Pederson, J. R., Neefus, C. D., Dawes, C. J., and Bray, T. L., 2008. Multiple assessments of introduced seaweeds in the Northwest Atlantic. ICES Journal of Marine Science, 65: 730-741.
Nelson, W., Heesch, S., Broom, J., Neill, K., Farr, T., and Dalen, T., 2007. Using molecular tools to tackle troublesome genera: Ulvaceae in New Zealand-diversity and introduced species revealed. Proceedings of the 22nd Annual Conference of the Australasian Society of Phycology and Aquatic Botany, 26–29 November 2007. Deakin University, Warrnambool, Victoria, 22.
Nyberg, C. D., and Wallentinus, I., 2005. Can species be used to predict marine macroalgal introductions? Biological Invasions, 7: 265-279.
Phillips, J. A., 1988. Field, anatomical and developmental studies on southern Australian species of Ulva (Ulvaceae, Chlorophyta). Australian Systematic Botany, 1: 411-456.
Rindi, F., Sartoni, G., and Cinelli, F., 2002. A floristic account of the benthic marine algae of Tuscany (Western Mediterranean Sea). Nova Hedwigia, 74 (1-2): 201-250.
Rindi, F., and Battelli, C., 2005. Spatio-temporal variability of intertidal algal assemblages of the Slovenian coast (Gulf of Trieste, northern Adriatic Sea). Botanica Marina, 48: 96-105.
Saunders, G. W., and Kucera, H., 2010. An evaluation of rbcL, tufA, UPA, LSU and ITS as DNA barcode markers for the marine green macroalgae. Cryptogamie, Algologie, 31 (4): 487-528.
Schneider, C. W., 2010. Report of a new invasive alga in the United States: ‘Heterosiphonia’ japonica in Rhode Island. Journal of Phycology, 46: 653-657.
Sfriso, A., 2010. Coexistence of Ulva rigida and Ulva laetevirens (Ulvales, Chlorophyta) in Venice Lagoon and other Italian transitional and marine environments. Botanica Marina, 53: 9-18.
Thornber, C. S., Guidone, M., and Deacutis, C., 2009. Community analyses of macroalgal blooms. Abstracts, 48thNortheast Algal Symposium, Univ. Massachusetts, Amherst, 37.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S., 2011. MEGA5: Molecular evolutionary genetics analysis using maximum-likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28 (10): 2731-2739.
(Edited by Qiu Yantao)
(Received July 17, 2013; revised September 22, 2013; accepted April 25, 2014)
© Ocean University of China, Science Press and Spring-Verlag Berlin Heidelberg 2014
* Corresponding author. E-mail: yxmao@ouc.edu.cn
杂志排行
Journal of Ocean University of China的其它文章
- Sedimentary Evolution of the Holocene Subaqueous Clinoform off the Southern Shandong Peninsula in the Western South Yellow Sea
- Identification of Fucans from Four Species of Sea Cucumber by High Temperature1H NMR
- Experimental Study on the Flow Around Two Tandem Cylinders with Unequal Diameters
- Revision of P-wave Velocity and Thickness of Hydrate Layer in Shenhu Area, South China Sea
- Study on Internal Waves Generated by Tidal Flow over Critical Topography
- Isolation and Characterization of Fucoidans from Five Brown Algae and Evaluation of Their Antioxidant Activity
