Development of a multiplex PCR assay to identify seven newcapsular gene loci of Streptococcus suis
2014-04-09LIUZhijieBAIXuemeiJIShaoboXUJianguoZHENGHan
LIU Zhi-jie,BAI Xue-mei,JI Shao-bo,XU Jian-guo,ZHENG Han
(Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, State KeyLaboratory for Infectious Disease Prevention and Control, National Institute for CommunicableDisease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing 102206, China)
Streptococcussuis(S.suis) is one of the most important swine pathogens worldwide, responsible for cases of septicemia with sudden death, meningitis, arthritis, endocarditis, and pneumonia amongst other diseases. It has significant impact on the swine industry[1]. Furthermore, it is also an emerging zoonotic agent. Humans can be infected when in close contact with pigs or pork products through skin wounds, or through consumption of raw pork[2-4]. During the past few years, the number of humanS.suisinfections reported worldwide has increased significantly, with the most cases reported in Asia[5-8].
Presently, 33 serotypes (type 1 through 31, 33, and 1/2) ofS.suishave been identified[9]. Thecpsclusters of 33 reference serotypes were published[10]. TheS.suisserotypes are routinely identified by the agglutination or co-agglutination tests using serotype-specific antisera[11]. However, these techniques are laborious, time-consuming, and relative expensive. In addition, auto-agglutinating strains cannot be serotyped using antisera. Molecular serotyping by PCR amplification of serotype specificcpsgenes does not require antisera and is an attractive alternative to compensate the existing agglutination and co-agglutination tests. We have developed three sets of multiplex PCR (mPCR) assays to identify all 33 serotypes ofS.suisbefore[12]. However, there are untypable strains using agglutinating by antisera remaining undetectable by our previous mPCR scheme.
To disclose the untypable reason of these strains using two methods described above, the genome of 14 field strains isolated from healthy pigs were sequenced by Solexa. Seven newcpsloci types were discovered. In the present study, we developed an mPCR assay targeting the serotype-specific polysaccharide polymerase genewzyto identify these newcpsloci types.
Material and methods
Bacterial strains and DNA templates preparation
FourteenS.suisfield isolates from healthy pigs were used for genome sequencing. Reference strains for 33S.suisserotypes, 1 to 31, 33, 1/2, andStreptococcusorisrattistrains originally classified as the reference strains forS.suisserotypes 32 (strain EA1172.91) and 34 (strain 92-2742) were from theS.suisstrain collection at the University of Montreal, Montreal, Canada[9]. A total of 91 field strains which cannot be serotyped using both agglutinating by antisera (serum provided by Statens Serum Institute, Copenhagen, Denmark) as well as the mPCR assays to identify known 33 serotypes[12]developed before isolated between 2011 and 2012 from clinically healthy pigs in slaughter houses in Beijing, Jiangsu Province and Sichuan Province were used for application of the mPCR assay. All isolates were serotyped using the agglutination test. The strains were grown overnight on Columbia blood base agar plates (Guangzhou Detgerm Microbiological Science, P. R. China) at 37℃ and a single colony was inoculated in 5 mL of Todd-Hewitt broth (THB, Oxoid Ltd., London, UK) and incubated for 8 hours at 37℃ in 5% CO2incubator.Streptococcuspneumoniastrains ATCC700657, ATCC700670, ATCC700676, ATCC700902, ATCC700906, ATCC49619,StreptococcusbovisATCC33317, andStreptococcuspyogenesATCC700294 were from our laboratory collection.Klebsiellapneumoniae46117-3,Streptococcuspyogenes32003,Streptococcussanguis32214,Enterococcusfaecalis32221,Streptococcusoralis32231,Streptococcuslutetiensis033,Streptococcusthermophilus20174,Streptococcusmutans10387,Streptococcusagalactiae10465, andStreptococcusacidominimus21026 were purchased from the China Center of Industrial Culture Collection. The template DNA was extracted using a Wizard genomic DNA purification kit (Promega, Madison, WI).
Whole genome sequencing and identification of the new cps locus
Genomic DNA was sequenced by Solexa sequencing after constructing a paired-end (PE) library with an average insert length of 500 bp to 2 000 bp. The reads were 100 bp in length generated with Illumina Solexa GA IIx (Illumina, San Diego, CA) and assembled into scaffolds using the program SOAP denovo (Release 2.04, http://soap.genomics.org.cn/soapdenovo.html). Eachcpslocus sequence was identified from the draft sequence based on theS.suiscpslocus characteristics previously reported[10,13-14]. The Artemis program (www.sanger.ac.uk) was used to identifycpsopen reading frames (ORFs) and annotations[15]. BLAST and PSI-BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) were used to search several databases[16]including the GenBank (www.ncbi.nlm.nih.gov/GenBank), the Clusters of Orthologous Groups (COG; www.ncbi.nlm.nih.gov/COG/), and Pfam (pfam.sanger.ac.uk) protein motif databases[17-18]. Thecpsgenes for each newcpstype were named withcpsfollowed by a letter from A to Z, in order.
Identification of serotype-specific genes in the cps loci
The local BLAST program BLAST+ applications (downloaded from ftp://ftp.ncbi.nlm.nih.gov/blast/executables/LATEST) were performed on a Microsoft Windows platform. The genome sequences of the 33 serotype reference strains published by our lab before and the 14 field strains sequenced in this study were used to build a local database. Eachcpsgene of sequenced strains in this study was compared and analyzed according to the method described in previous study[12].
Sequence alignment and comparisons were performed using the ClustalW program[19]. The phylogenetic trees for thewzygene of the newcpsloci and the reference strains were generated by the neighbor-joining method using the program MEGA[21].
Primer design and conditions of mPCR reaction
Using the Primer-BLAST program (http://www.ncbi.nlm.nih.gov/tools/primer-blast/), primers were designed to have similar physical characteristics in order to allow simultaneous amplification in the same conditions and multiplex reactions. The lengths of the primers were between 22 and 24 bp, their melting temperatures were between 49.9 and 50.2 ℃, and the expected amplicon sizes ranged between 254 and 744 bp. The internal control primers ofthrAwere used as described before[12]. The primers used for the mPCR assay are shown in Table 1. The primers were synthesized by Sangon Biotech (Shanghai) and dissolved in TE buffer (10 mM Tris-Cl, 1 mM EDTA) to obtain 20 μM stock solutions.
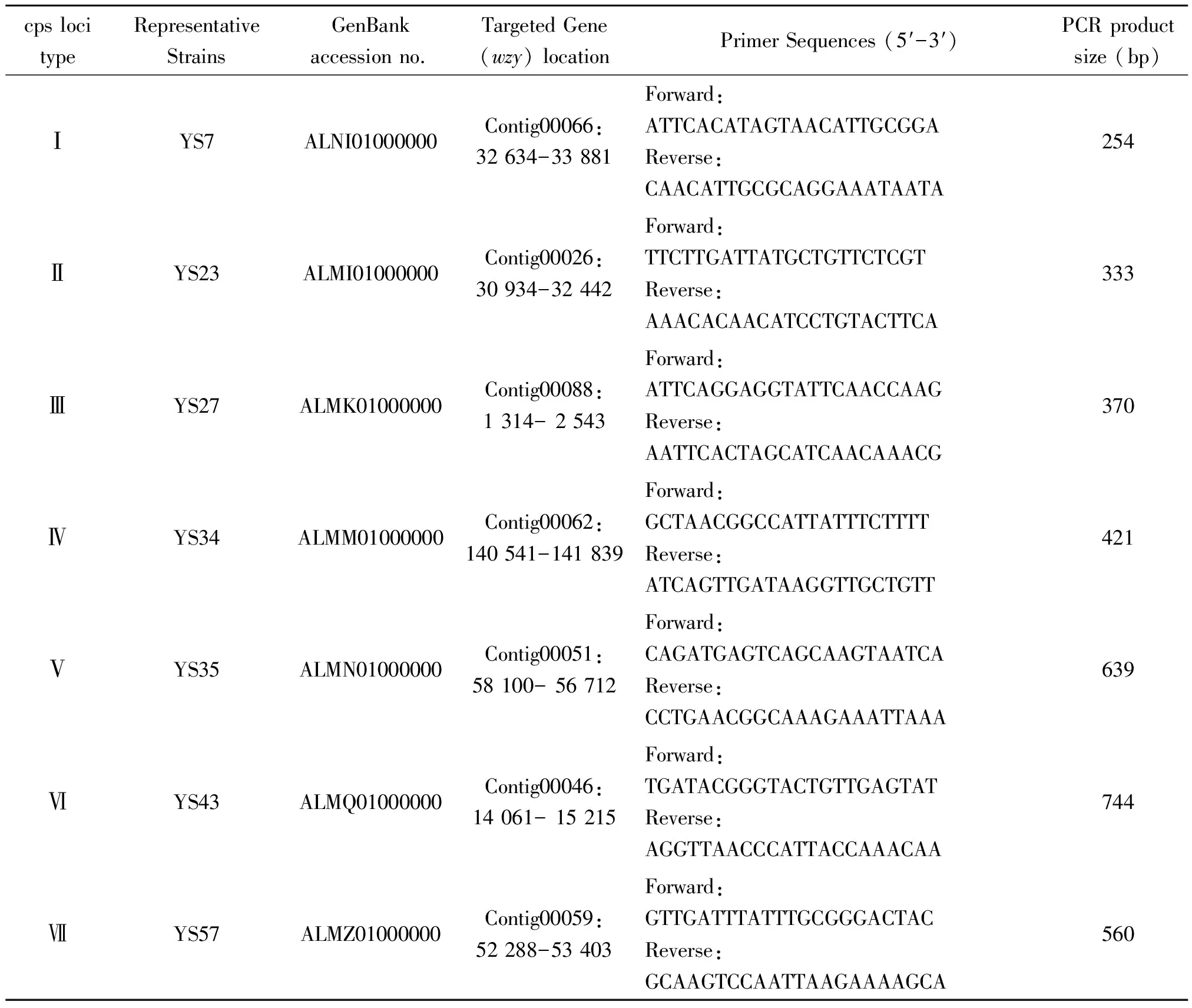
Tab.1 New cps loci type specific primers used in this study
mPCR was performed using 2×TaqPCR Master Mix containingTaqDNA polymerase: 0.05 units/μL; MgCl2: 4 mM; dNTP: 4 mM; and buffer (CoWin Biotech Co., Ltd, Beijing, China). The reaction mixture (20 μL) for each PCR consisted of 10 μL 2×TaqPCR Master Mix, and 0.2 μM of each primer. The PCR program for the mPCR reactions was as follows: 94 ℃ for 5 min, followed by 30 cycles: 94 ℃ for 30 sec, 58 ℃ for 40 sec, and 72 ℃ for 50 sec; with a final extension of 72 ℃ for 5 min in a thermocycler (Senso, Germany). The PCR products were analyzed with gel electrophoresis using 2% agarose gels.
Sensitivity, specificity and application of the mPCR assay
The detection sensitivity for genomic DNA was determined. Genomic DNA was 10-fold serially diluted to concentrations of 10 ng, 1 ng, 100 pg, 10 pg, 1 pg, and 100 fg/μL. One microliter of each dilution was used as the DNA template. Genomic DNA of 33 serotype referenceS.suisstrains, 19 otherStreptococcusspp. strains and oneKlebsiellapneumoniastrain were used to detect the specificity of the mPCR assay.
The mPCR assay was used to detect 91 untypableS.suisfield strains, which can not be serotyped using both agglutinating by antisera as well as the mPCR assays to identify known 33 serotypes developed before[12]. The fullcpsof each detectable strain using this mPCR assay was amplified by walking PCR according the correspondingcpsloci type. The DNA products were sequenced, and the sequences were compared with the representative newcpsloci.
Results
Identifying new cps loci and target genes for the mPCR assay
Among 14 sequencedS.suisstrains, we discovered 7 newcpsloci types. The length of the newcpsgene clusters ranged from 16 708 bp to 33 158 bp. Percentage G + C content of all the newcpsgene clusters (33.6-36.7%) was lower than those of the several reportedS.suisgenomes (41.0-41.4%) (Table 2). The characteristic of newcpsloci was similar to the 33 referencecpsgene clusters[10]in that (Ⅰ) the chromosomal loci of all the 7 newcpsclusters were classified into pattern I-b, flanked by theorfZ-orfXregion and theglfgene (UDP-galactopyranose mutase gene; corresponding to SSU0563 in P1/7); (Ⅱ) the first four genescpsA-Din thecpscluster were conserved, and type-specific genes were located in either the central region or the last parts of thecpsgene clusters; (Ⅲ) type-specificwzyexisted in all of the newcpstypes. Four to 8 type specific genes were identified in each newcpsloci type (Table 2). The newcpstype specific genes encoded glycosyltransferase, acetyltransferase, phosphotransferase, nucleotidyltransferase, phosphate dehydrogenase, polysaccharide polymerase (Wzy), or flippase (Wzx). Moreover, high sequence divergence was recognized in thewzygenes of the strains between newcpsclusters and reference serotypes inS.suis(Figure 1). We proposed these were newcpsloci types because of the type specific genes in newcpsloci were very different from the knowncpsgenes.
Thewzygene proved to be an ideal target gene to develop mPCR assays to discriminate the 33 serotypes previously[12]. Therefore, thewzygene of newcpsloci was chosen as the target gene to develop the mPCR assay to identify the newcpsloci type in this study.
The 7 newcpsloci types named as Ⅰ, Ⅱ, Ⅲ, Ⅳ, Ⅴ, Ⅵ and Ⅶ. Thecpsof 161_00P5 (GenBank No. ALKX01000000), YS7 (GenBank No. ALNI01000000), YS21 (GenBank No. ALMH01000000), YS49 (GenBank No. ALMT01000000), YS56 (GenBank No. ALMY01000000) were assigned to type Ⅰ. Thecpsof YS23 (GenBank No. ALMI01000000) and YS72 (GenBank No. ALNF01000000) were assigned to type Ⅱ. Thecpsof YS27 (GenBankNo. ALMK01000000) and YS74 (GenBank No. ALNG01000000) were assigned to type Ⅲ. Thecpsof YS34 (GenBank No. ALMM01000000), YS35 (GenBank No. ALMN01000000) and YS43 (GenBank No. ALMQ01000000) were assigned to type Ⅳ, Ⅴ and Ⅵ respectively. Thecpsof YS46 (GenBank No. ALMS01000000) and YS57 (GenBank No. ALMZ01000000) were assigned to type Ⅶ. We chose one representative strain (Table 1) for each newcpstype to develop the mPCR assay.

Fig.1Thetreewasconstructedusingtheneighbor-joiningalgorithmbasedonwzygenesofthe7representativestrainswithnewcpslocitype(inbluecolor)and33referencestrainsofS.suis(Bar,sequencedissimilarity)
Development of the mPCR assay
First, we designed type-specific PCR primers based on thewzygene and performed simplex PCRs to determine the specificity of each primer pair using template DNA extracted from the 33 reference strains and the representative strains of newcpstypes. Each pair of primers amplified the predicted PCR product specifically from the DNA samples.
The mPCR assay was then designed based on the simplex PCR assays above. A primer pair that amplified a 120 bp fragment from thethrAgene was added to mPCR as an internal control. DNA samples prepared from the representative strains of newcpstypes were analyzed using the mPCR assays. For each DNA sample, two bands were pro-duced, one of which was the internal control, as expected, while the other was thecpstype-specificwzygene. No cross-amplification product was detected from these strains. The amplicon sizes allowed good separation on 2% agarose gels, where each PCR product could be unambiguously identified by size (Figure 2).


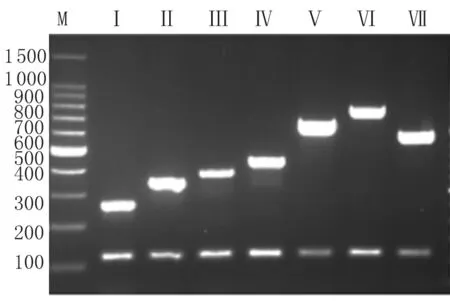
Fig.2MultiplexPCRproductsof7S.suisrepresentativestrainswithnewcpslocitype
PCR products were electrophoresed on a 2% (wt/vol) agarose gel, stained with goldenview, and photographed under UV light. Thecpsloci types are indicated above the lanes.
Lane M: 100-bp DNA ladder markers (Takara, Dalian, China), the sizes (bp) are indicated on the left.
Sensitivity, specificity and application of the mPCR assay
The detection limit of the mPCR assay for the representative strain of each newcpsloci type was 100 pg. Three independent experiments were performed to establish the sensitivity of the mPCR assay. The specificity of the mPCR assay was tested using the 33 serotype-referenceS.suisstrains, 19 otherStreptococcusspp. strains and oneKlebsiellapneumoniastrain. Non-specific amplification band was not observed in any of the samples tested (data not shown).
To determine the distribution of the newcpsloci, the mPCR assay was conducted in 91S.suisuntypable strains. Seventy-one of them were positive and 20 strains cannot be detected by the mPCR assay. There were 34 isolates belong tocpstypeⅠ, 14 isolates belong tocpstypeⅡ, 11 isolates belong tocpstype Ⅲ, 8 isolates belong tocpstype Ⅳ, 1 isolate belongs tocpstype Ⅴ, and 3 isolates belong tocpstype Ⅶ. Except for the representative strain YS43, there was no isolate detected ascpstype Ⅵ (Table 3).
Tab.3ApplicationofthemPCRassayin91S.suisstrains
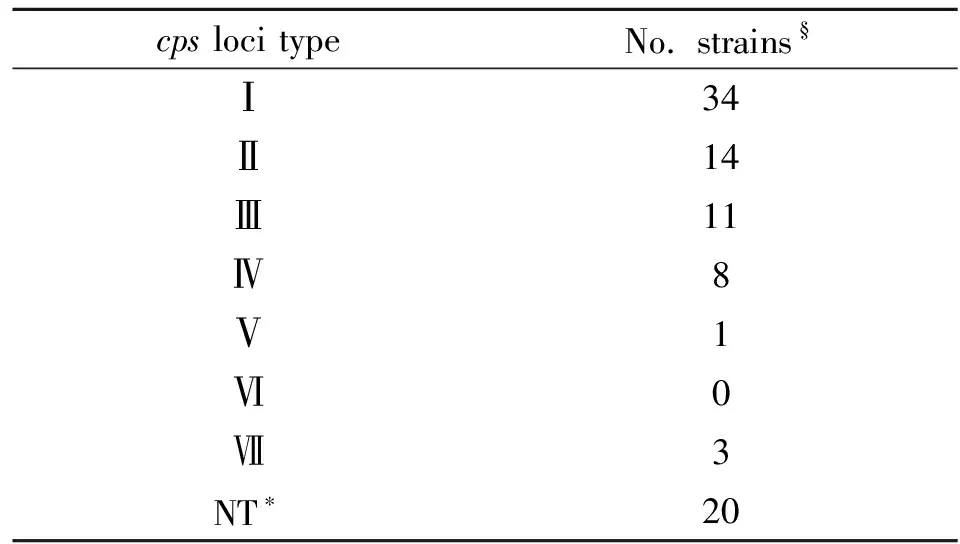
cps loci typeNo. strains§Ⅰ34Ⅱ14Ⅲ11Ⅳ8Ⅴ1Ⅵ0Ⅶ3NT∗20
Note:§The 91S.suisstrains used here which were untypable strains using both agglutinating by antisera as well as the mPCR assays to identify known 33 serotypes developed before.
*NT represents mPCR assay negative strains in this study.
Discussion
Serotyping is one of the most important diagnostic tools forS.suisand remains a valuable method to understand the epidemiology of a particular outbreak, monitor serotype prevalence, as well as guide vaccine development. TheS.suisserotypes are determined by the antigenicity of the capsule[21-23]. The type specific genes in thecpsloci are the basis ofS.suismolecular serotyping[12,24-25].
In this study, we used whole genome sequencing to obtain the fullcpsgene cluster from 14 untypable strains, and discovered 7 newcpsclusters. Comparing to the genome database we built, several type specific genes were identified for each newcpsloci type, including the polysaccharide polymerase genewzy. The same results were found in previous study[12], the polysaccharide polymerase genewzywas in newcpsloci, one of the type-specific genes shared very little DNA sequence identity with 33 reference strains and the other newcpsclusters (Figure 1). Therefore, it is ideally suited as a target for developing the mPCR assay to identify these newcpsclusters.
To our knowledge, this is the first study to state the 7 newcpsloci types inS.suisand we also developed a molecular protocol for detection in one mPCR mixture reaction. Among 91 detected strains, 71 were unambiguously proved as positive strains. The previous studies[10,12,24-25]indicate thecpslocus is usually conserved among different strains belonging to the same serotype or the samecpsloci type. To confirm the reliability of the mPCR assay, the full-lengthcpsof each mPCR positive strain was sequenced. The high sequence similarity among strains harboring the samewzygene was found (data not shown). Thus, the mPCR assay is reliable and practical to identify the 7 newcpstypes simultaneously. Whether these strains belong to new serotypes will be deciphered through immunizing animals in further research. There are 20 strains still remaining undetectable. We cannot rule out the possibility that these strains possess unknowncpsloci.
In conclusion, the mPCR assay we developed is rapid, specific and could successfully validate protocol to identify the 7 newcpstypes. This assay may serve as an effective way for screening the untypableS.suisstrains, and may help us to uncover other novelcpsclusters and new serotypes.
[1]Wertheim HF, Nghia HD, Taylor W, et al.Streptococcussuis: an emerging human pathogen[J]. Clin Infect Dis, 2009, 48(5): 617-625. DOI: 10.1086/596763
[2]Kay R, Cheng AF, Tse CY.Streptococcussuisinfection in Hong Kong[J]. QJM, 1995, 88(1): 39-47.
[3]Yu HJ, Liu XC, Wang SW, et al. Matched case-control study for risk factors of humanStreptococcussuisinfection in Sichuan Province, China[J]. Chin J Epidemiol, 2005, 26(9): 636-639. (in Chinese)
[4]Takeuchi D, Kerdsin A, Pienpringam A, et al. Population-based study ofStreptococcussuisinfection in humans in Phayao Pro-vince in northern Thailand[J]. PLoS One, 2012, 7(2): e31265. DOI: 10.1371/journal.pone.0031265
[5]Huang YT, Teng LJ, Ho SW, et al.Streptococcussuisinfection[J]. J Microbiol Immunol Infect, 2005, 38(5): 306-313.
[6]Hui AC, Ng KC, Tong PY, et al. Bacterial meningitis in Hong Kong: 10-years' experience[J]. Clin Neurol Neurosurg, 2005, 107(5): 366-370. DOI: 10.1016/j.clineuro.2004.10.006
[7]Yu H, Jing H, Chen Z, et al. HumanStreptococcussuisoutbreak, Sichuan, China[J]. Emerg Infect Dis, 2006, 12(6): 914-920. DOI: 10.3201/eid1206.051194
[8]Mai NT, Hoa NT, Nga TV, et al.Streptococcussuismeningitis in adults in Vietnam[J]. Clin Infect Dis, 2008, 46(5): 659-667. DOI: 10.1086/527385
[9]Hill JE, Gottschalk M, Brousseau R, et al. Biochemical analysis, cpn60 and 16S rDNA sequence data indicate thatStreptococcussuisserotypes 32 and 34, isolated from pigs, areStreptococcusorisratti[J]. Vet Microbiol, 2005, 107(1): 63-69.
[10]Okura M, Takamatsu D, Maruyama F, et al. Genetic analysis of capsular polysaccharide synthesis gene clusters from all serotypes ofStreptococcussuis: potential mechanisms for generation of capsular variation[J]. Appl Environ Microbiol, 2013, 79(8): 2796-2806. DOI: 10.1128/AEM.03742-12
[11]Higgins R, Gottschalk M. An update onStreptococcussuisidentification[J]. J Vet Diagn Invest, 1990, 2(3): 249-252.
[12]Liu Z, Zheng H, Gottschalk M, et al. Development of multiplex PCR assays for the identification of the 33 serotypes ofStreptococcussuis[J]. PLoS One, 2013, 8(8): e72070. DOI: 10.1371/journal.pone.0072070
[13]Smith HE, Damman M, van der Velde J, et al. Identification and characterization of the cps locus ofStreptococcussuisserotype 2: the capsule protects against phagocytosis and is an important virulence factor[J]. Infect Immun, 1999, 67(4): 1750-1756.
[14]Wang K, Fan W, Cai L, et al. Genetic analysis of the capsular polysaccharide synthesis locus in 15Streptococcussuisserotypes[J]. FEMS Microbiol Lett, 2011, 324(2): 117-124. DOI: 10.1111/j.1574-6968.2011.02394.x
[15]Mural RJ. ARTEMIS: a tool for displaying and annotating DNA sequence[J]. Brief Bioinform, 2000, 1(2): 199-200. DOI: 10.1093/bib/1.2.199
[16]Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs[J]. Nucleic Acids Res, 1997, 25(17): 3389-3402.
[17]Tatusov RL, Natale DA, Garkavtsev IV, et al. The COG database: new developments in phylogenetic classification of proteins from complete genomes[J]. Nucleic Acids Res, 2001, 29(1): 22-28.
[18]Bateman A, Birney E, Cerruti L, et al. The Pfam protein families database[J]. Nucleic Acids Res, 2002, 30(1): 276-280.
[19]Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice[J]. Nucleic Acids Res, 1994, 22(22): 4673-4680.
[20]Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods[J]. Mol Biol Evol, 2011, 28(10): 2731-2739. DOI: 10.1093/molbev/msr121
[21]Demoor CE. Septicaemic infections in pigs, caused by haemolytic streptococci of new Lancefield groups designated R, S, and T[J]. Antonie Van Leeuwenhoek, 1963, 29(0): 272-280.
[22]Elliott SD. Streptococcal infection in young pigs. I. An immunochemical study of the causative agent (PM streptococcus)[J]. J Hyg (Lond), 1966, 64(2): 205-212.
[23]Windsor RS, Elliott SD. Streptococcal infection in young pigs. IV. An outbreak of streptococcal meningitis in weaned pigs[J]. J Hyg (Lond), 1975, 75(1): 69-78.
[24]Kerdsin A, Dejsirilert S, Akeda Y, et al. FifteenStreptococcussuisserotypes identified by multiplex PCR[J]. J Med Microbio, 2012, 61(12): 1669-1672. DOI: 10.1099/jmm.0.048587-0
[25]Wang K, Sun X, Lu C. Development of rapid serotype-specific PCR assays for eight serotypes ofStreptococcussuis[J]. J Clin Microbiol, 2012, 50(10): 3329-3334. DOI: 10.1128/JCM.01584-12
