Aberrant DNA methyltransferase 1 expression in clear cell renal cell carcinoma development and progression
2014-03-21MingLiYingWangYongshengSongRengeBuBoYinXiangFeiQizhenGuoBinWu
Ming Li, Ying Wang, Yongsheng Song, Renge Bu, Bo Yin, Xiang Fei, Qizhen Guo, Bin Wu
1Department of Urology, Shengjing Hospital of China Medical University, Shenyang 110004, China;2Department of Cell Biology, Harvard Medical School, Boston, MA, 02115, USA;3Department of Nuclear Medicine, The First Affiliated Hospital of China Medical University, Shenyang 110001, China
Correspondence to: Bin Wu. Department of Urology, Shengjing Hospital of China Medical University, Shenyang 110004, China. Email: wubin_cmu@163.com.
Aberrant DNA methyltransferase 1 expression in clear cell renal cell carcinoma development and progression
Ming Li1,2, Ying Wang3, Yongsheng Song1, Renge Bu1, Bo Yin1, Xiang Fei1, Qizhen Guo1, Bin Wu1
1Department of Urology, Shengjing Hospital of China Medical University, Shenyang 110004, China;2Department of Cell Biology, Harvard Medical School, Boston, MA, 02115, USA;3Department of Nuclear Medicine, The First Affiliated Hospital of China Medical University, Shenyang 110001, China
Correspondence to: Bin Wu. Department of Urology, Shengjing Hospital of China Medical University, Shenyang 110004, China. Email: wubin_cmu@163.com.
Objective:To better understand the contribution of dysregulated DNA methyltransferase 1 (DNMT1) expression to the progression and biology of clear cell renal cell carcinoma (ccRCC).
Methods:We examined the differences in the expression of DNMT1 in 89 ccRCC and 22 normal tissue samples by immunohistochemistry. In addition, changes in cell viability, apoptosis, colony formation and invading ability of ccRCC cell lines (786-0 and Caki-1) were assessed after transfection with DNMT1 siRNA.
Results:We found DNMT1 protein was signifcantly higher expressed in ccRCC than that of in no-tumor tissues (56.2% and 27.3%, respectively, P=0.018). The expression of DNMT1 was strongly associated with ccRCC tumor size, tumor pathology stage, histological grading, lymph node metastasis, vascular invasion, recurrence and prognosis. Moreover, knockdown of DNMT1 expression signifcantly inhibited ccRCC cell viability, induced apoptosis, decreased colony formation and invading ability.
Conclusions:Expression of DNMT1 protein is increased in ccRCC tissues, and DNMT1 expression is associated with poor prognosis of patients. Experiments in vitro further showed DNMT1 played an essential role in proliferation and invasion of renal cancer cells. Moreover, targeting this enzyme could be a promising strategy for treating ccRCC, as evidenced by inhibited cell viability, increased apoptosis, decreased colony formation and invading ability.
Clear cell renal cell carcinoma (ccRCC); DNA methyltransferase 1 (DNMT1); immunohistochemistry; siRNA
View this article at:http://dx.doi.org/10.3978/j.issn.1000-9604.2014.08.03
Introduction
Renal cell carcinoma (RCC) is the most lethal urologic tumor and the sixth leading cause of cancer deaths in Western countries. Each year, around 200,000 patients are diagnosed with this malignancy resulting in approximately 100,000 deaths, and its incidence is increasing steadily in recent years (1). Nearly 25-30% of patients with RCC have evidence of metastases at initial presentation (2). RCC is represented by 80% by clear cell renal cell carcinoma (ccRCC), originating from the renal proximal tubule (3). ccRCC is resistant to both radiation and chemotherapy, and the prognosis remains poor. Although standard pathological features such as tumor grade and stage can be used to decide on treatments, to explore novel molecular markers, which could be served as prognostic factors and therapeutic targets for ccRCC is still an important issue.
During carcinogenetic development, epigenetic changes occur at least as frequently as genetic mutations and deletions. Aberrant DNA methylation plays a key role in carcinogenesis, leading to the epigenetic silencing of the expression of tumor-suppressor genes involved in cell cycle regulation, apoptosis, and DNA repair (4,5). In such cases, DNA methylation is typically mediated by DNA methyltransferases, particularly DNA methyltransferase1 (DNMT1), DNMT3A, and DNMT3B (6). DNMT1 is signifcantly overexpressed in several tumor types (7,8). In a variety of cancers, DNMT1, DNMT3A, and DNMT3B were reported to be highly expressed and associated with poor prognosis, including breast, liver, lung, colorectum, stomach, bladder, prostate, cervical cancers, lymphomas and other malignancies (9-20). Until now, there is minimal amount of report on DNMT1 in RCC. A report in 2006 (21) found that the incidence of nuclear immunoreactivity for DNMT1 tended to be higher in proximal tubules from non-tumorous renal tissue than in those from normal renal tissue, and was significantly higher in RCCs, however, there have been no reports regarding biological behavior alterations of ccRCC cells after knockdown of DMNT1 expression and the underlying molecular events in vitro. Thus, in this study, we analyzed the expression of DNMT1 protein in ccRCC tissue specimens and investigated their association with clinicopathological features and patient survival. Then, we knocked down DNMT1 expression using siRNA to assess the effects on the regulation of biological behavior of ccRCC cells and the underlying molecular mechanisms.
Materials and methods
Patients and tissue samples
ccRCC tissue was collected from radical nephrectomy specimens performed between January 2004 and January 2012 at Department of Urology, Shengjing Hospital of China Medical University. The tumor cases included 89 cases with histologically confirmed ccRCC and 22 cases of adjacent non-tumor tissues. The criteria for study enrollment were as follows: patients with histopathologically diagnosed ccRCC who were newly diagnosed, untreated without a history of other tumors, and subsequently underwent radical nephrectomy. Histological diagnosis was established according to the guidelines of the World Health Organization (22). Cases were selected according to tissue availability and were not stratified for any known preoperative or prognostic factor. None of the patients underwent chemotherapy, radiotherapy, or adjuvant treatment before surgery. We obtained the written informed consent from all the patients. The Institute Research Medical Ethics Committee of Shengjing Hospital of China Medical University approved the research protocol. The patients were carefully followed up by consulting their case documents and through telephone monitoring.
Immunohistochemistry
Formalin-fxed and paraffn-embedded tissue samples were cut into 4-mm thick sections and mounted onto poly-L-lysine-coated glass slides. For immunohistochemical staining, the sections were deparaffinized in xylene, rehydrated in a series of alcohol, and washed in the tap water. The sections were then cooked in 10 mm sodium citrate buffer, pH 6.0, for 10 min in an autoclave for antigen retrieval. Endogenous peroxidase activity was blocked by incubating the sections in 3% H2O2at 37 ℃ for 20 min. After that, the sections were blocked to avoid nonspecific binding by addition of a 10% normal goat serum at 37 ℃for 30 min and then incubated for 4 ℃ overnight with the polyclonal antibody against DNMT1 (sc-20701, 1:250 dilution Santa Cruz Biotechnology, USA). The specificity of antibodies had been confirmed by using Western blot analysis (data not shown). In the next day, the sections were washed five times with 0.01 mol/L phosphate-buffered saline (PBS; pH 7.4) for 15 min and then incubated with a biotinylated secondary antibody for 30 min at 37 ℃ in the dark. After that, the sections were incubated with a streptavidin horseradish peroxidase solution for another 30 min (LSAB kit; Dako, Glostrup, Denmark), washed in PBS, and stained with DAB (3, 3-diaminobenzidine). Finally, the sections were counterstained with Mayer’s hematoxylin, dehydrated, and mounted. Negative controls were run in parallel, and were generated by PBS replacing the anti-DNMT1 antibody.
The immunostained sections were evaluated by two investigators who were blinded to the patients’clinicopathological characteristics. For each slide, the number of DNMT1 positive cells was counted in ten felds at ×200 magnifications, and the percentage of positively stained cells was determined. The percentage of positively stained cells was graded semi-quantitatively according to a four-point scoring system as follows: negative (–), 0; weakly positive (+), <25%; moderately positive (++), 26-50%; and strongly positive (+++), >50%.
ccRCC cell line and culture
786-0 human ccRCC cell line was obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in RPMI 1,640 supplemented with 10% FCS. Caki-1 human ccRCC cell line was obtained from Tiancheng Technology Co. Ltd (Shanghai, China) and cultured in DMEM supplemented with 10% FCS. Cellswere harvested when they were in the logarithmic phase of growth for use in the following experiments.
RNA isolation and quantitative-reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was isolated from transfected cells as well as untransfected cells after transfection, using a trizol reagent (Invitrogen) and was then reversely transcribed into cDNA using a PrimeScriptTM RT kit (TakaRa, Dalian, China). The expression of DNMT1 was detected by qPCR in ABI7500 (Applied Biosystems, Foster City, CA, USA). The primer sequences were 5'-GTTCTTCCTCCTGGAGAATGTCA-3' and 5'-GGGCCACGCCGTACTG-3' for DNMT1.The program had an initial denaturation at 95 ℃ for 1 min followed by 40 cycles of 95 ℃ for 15 s and 60 ℃ for 15 s. The relative quantity (RQ) was determined using the 2(–ΔΔCt) method, a β-actin primer was used as a loading control, and each experiment was repeated six times.
Construction of DNMT siRNA vector and gene transfection
The DNMT1 specifc siRNA was chosen from the GenBank sequences (accession #NM_001379.1) and synthesized by Ambion Inc. (Austin, TX, USA). The DNMT1 siRNA sequences were 5'-GGAUGAGAAGAGACGUAGAtt-3' and 5'-UCUACGUCUCUUCUCAUCCtg-3'. Negative control siRNA was also purchased from Ambion Inc. (Catalog #4,390,843). Final concentrations were 7 nM. The OptiMEM (Gibco BRL Inc. USA) and Lipofectamine 2,000 Transfection Kits (Invitrogen) were used for siRNA transfections. Cells were plated at 1×105cells per well in culture dishes for overnight growth and transfected by siRNA on the following day. Six hours after transfection, the medium was removed and replaced with RPMI 1,640 or DMEM, and the cells were allowed to grow for 48 h. Then, viability, apoptosis, colony formation and invading ability were assayed.
Protein extraction and western blot
Total cellular protein was extracted from cells using an M-PER mammalian protein extraction buffer (Pierce, Rockford, IL) containing 0.5 mm PMSF. The protein samples were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE, 12%) and electronically transferred onto PVDF membranes (Millipore, Billerica, MA, USA). The membranes were incubated with an anti-DNMT1 antibody (sc-20701, 1:250 dilution Santa Cruz Biotechnology, USA) or an anti-βactin (1:5000, Sigma, St Louis, MO, USA) antibody at 4 ℃ overnight, followed by a secondary antibody for 2 h at room temperature. The protein bands were detected with enhanced chemiluminescence (ECL) reagent (Invitrogen).
Cell viability Tetrazolium Salt-8 (WST-8) assay
Briefly, cells were transfected with DNMT1 or negative control siRNA oligonucleotides. Seventy-two hours later, 10 µL of WST-8 reagent was added to each well and incubated for an additional 4 h. The absorbance rate was then measured at 450 nm with an ELISA plate reader (Thermo ELISA Reader, USA).
Flow cytometry assay
Apoptosis levels were detected using fow cytometry (Becton Dickinson) with an Annexin V-FITC Kit (Jingmei Biotech Company, Shanghai, China) according to the manufacturer’s protocol.
In vitro colony formation assay
Cells were suspended in 0.1 mL of culture medium with 10% FBS, and 1,000 cells were plated in culture dishes with 1 mL of methylcellulose-containing culture medium supplemented with 15% FBS. The number of colonies was counted on day 14. Colonies were stained and counted by applying diff-quick staining kit (Siemens, Munich, Germany).
Matrigel invasion assay
Invasion of tumor cells into Matrigel was examined with a BD BioCoat Matrigel Invasion Chamber (BD Biosciences). Cells were seeded in culture medium without FCS in the Matrigel invasion upper chamber and cultured for 72 h. The lower chamber contained culture medium with 10% FBS. Invading cells were stained with a diff-quick staining kit (Siemens, Munich, Germany). The number of invading cells was counted in four microscopic fields per well at a magnification of 20, and the extent of invasion was expressed as the average number of cells per square millimeter.
Statistical analysis
Statistical analysis was performed with SPSS version 19 (SPSS Inc., Chicago, IL, USA). Comparison of DNMT1 expression between samples and the difference of the variables (such as expression of DNMT1 mRNA vs. tumor cell viability, apoptosis, colony formation and invading ability) between the differential siRNA transfected groups, were analyzed with a Mann-Whitney U test. Chi-square tests were applied to assess associations between expression of DNMT1 and clinicopathological parameters. Univariate survival analysis was performed with a Kaplan-Meier logrank test, while multivariate survival analysis was performed with a Cox Regression test. A two-tailed P<0.05 was considered statistically signifcant.
Results
Patient characteristics
The clinicopathological data from the patients are shown in Table 1. The mean age of the patients at surgery was 53 years (rang, 15-84 years), and 50 (56.2%) of the patients were diagnosed before 55 years old. Fifty-four (60.7%) were male. Clinical follow-up data, as annually assessed survival time was available for all patients. The median follow-up time of all cases was 49.5 months, ranging from 14 to 118 months. Twenty-fve (28.1%) patients exhibited recurrence and 21 (23.6%) patients died from ccRCC during follow up. The pT status was as follows: pT1 and pT2-67 (75.3%), pT3 and pT4-22 (24.7%). Thirteen (14.6%) patients had pathologically confrmed nodal metastases. Fifty-fve (61.8%) patients had no nodal metastases (pN0). In 21 (23.6%) patients lymph nodes were not examined (pNx). Tumor grades, according to Fuhrman, were G1-23 (25.8%), G2-30 (33.7%), G3-24 (27.0%) and G4-12 (13.5%), respectively.
Expression of DNMT1 in ccRCC and no-tumor tissues
The expression of DNMT1 protein in clear cell renal cell cancer and no-tumor tissues is summarized in Table 2. In our study, DNMT1 protein was significantly highly expressed in clear cell renal cell cancer tissues than that of no-tumor tissues (Mann-Whitney U-test, P=0.018). Briefly, the positive rates for DNMT1 expression in the ccRCC tissues were 56.2%, which was significantly higher than those of no-tumor tissues (27.3%). Representative expression patterns of immunohistochemical staining of DNMT1 in ccRCC and non-tumor tissues were shown in Figure 1.
Association of DNMT1 expression with clinicopathological characteristics
The correlation analysis of DNMT1 with clinicopathological factors in ccRCC was shown in Table 3. Our data showed that DNMT1 expression was significantly associated with tumor size (P=0.040), tumor pathology stage (P=0.022), histopathological grading (P=0.004), lymph node metastasis (P=0.029) and vascular invasion (P=0.037). However, there were no association between DNMT1 expression with gender (P=0.772), age (P=0.695), or tobacco smoking (P=0.379).
Association of DNMT1 expression with survival of the patientsThe correlation between DNMT1 expression andprognosis in ccRCC patients was analyzed with the Kaplan-Meier method. We observed that the expression of DNMT1 in ccRCC was significantly correlated with overall survival (OS) and disease free survival (DFS) (P=0.002 and 0.026, respectively, Table 4). The log-rank test further demonstrated that the OS and DFS time were both signifcantly different between groups with and without expression of DNMT1, which indicated expression of DNMT1 was correlated with a shorter survival time (Figure 2). Other clinicopathologic parameters, including tumor size (P=0.003 and 0.015, respectively), tumor pathology stage (P=0.002 and 0.006, respectively), histological grade (P=0.003 and 0.022, respectively), lymph node metastasis (P=0.024 and 0.036, respectively), and vascular invasion (P=0.007 and 0.003, respectively) were also significantly correlated with OS and disease-free survival in univariate analysis (Table 4). In addition, multivariate analysis using the Cox proportional hazards model showed that the expression of DNMT1 was an independentprognostic factor for OS in patients with ccRCC (P=0.031), the traditional tumor size, tumor pathology stage, histological grading, lymph node metastasis, and vascular invasion were independent predictors of both OS and disease-free survival (all P<0.05) (Table 5).
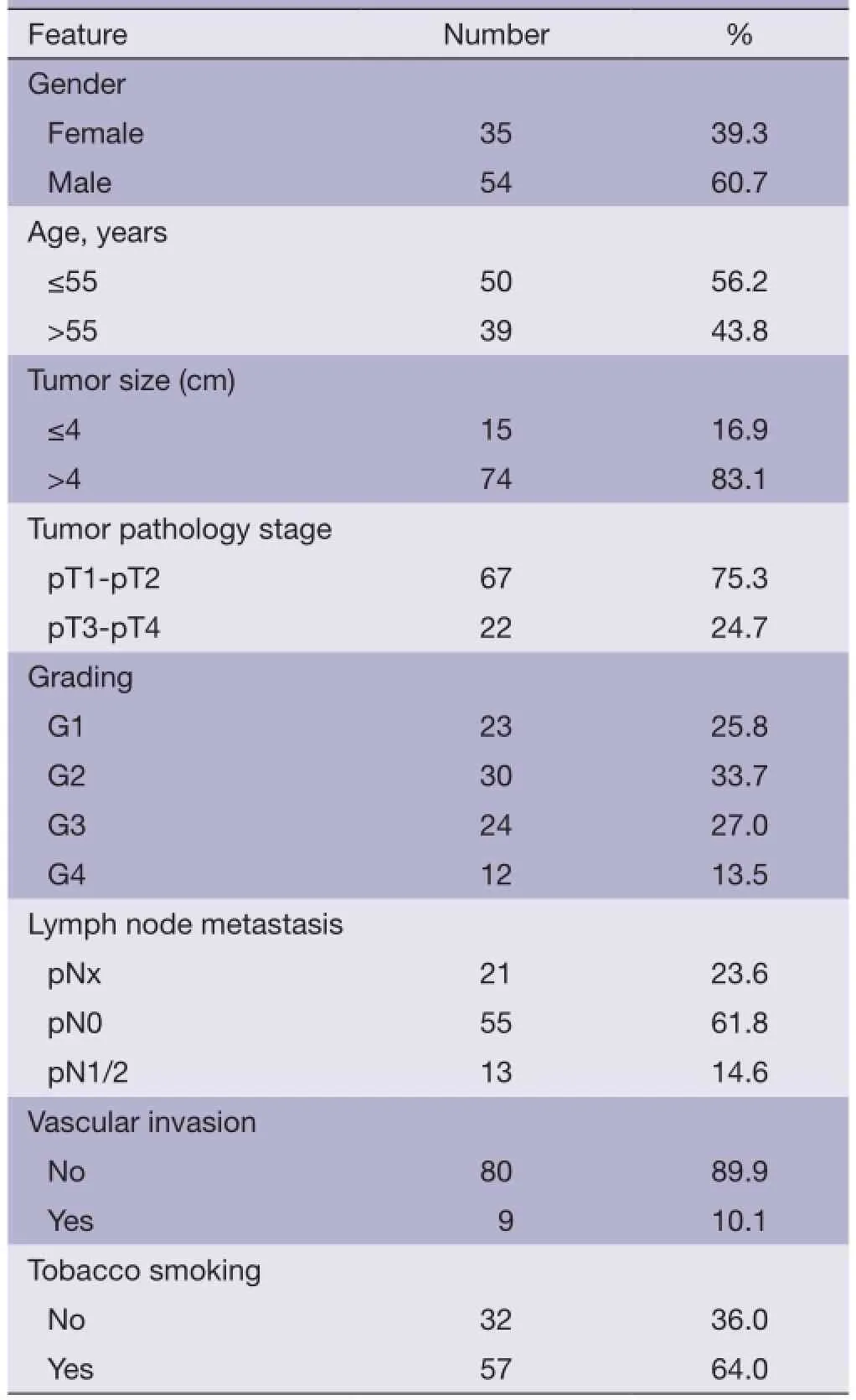
Table 1 Patient characteristics

Table 2 DNMT1 expression in ccRCC and non-tumor tissues
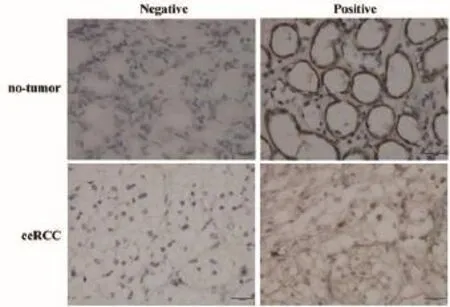
Figure 1 Nuclear immunostaining of DNMT1 protein in notumor and ccRCC tissues (Original magnifcation: ×400). ccRCC, clear cell renal cell carcinoma; DNMT1, DNA methyltransferase 1.

Table 3 Correlation between DNMT1 expression and clinicopathological factors of ccRCC
Effects of DNMT1 silencing on cell viability, apoptosis, colony formation and invading ability in ccRCC cell line
The DNMT1 were knocked down in ccRCC cell lines 786-0 and Caki-1. As shown in Figure 3, qRT-PCR and Western blot data showed that transfection of DNMT1 siRNA significantly reduced expression of their corresponding mRNA and protein. We found that transfection of DNMT1 siRNA significantly inhibited the viability of ccRCC cells and induced apoptosis (Figure 4A,B).Then, the ability for colony formation and invasion in vitro was evaluated. As shown in Figure 4C,D, when DNMT1 was inhibited by siRNA, clonies and invasive cell number reduced in 786-0 and Caki-1 cell lines.
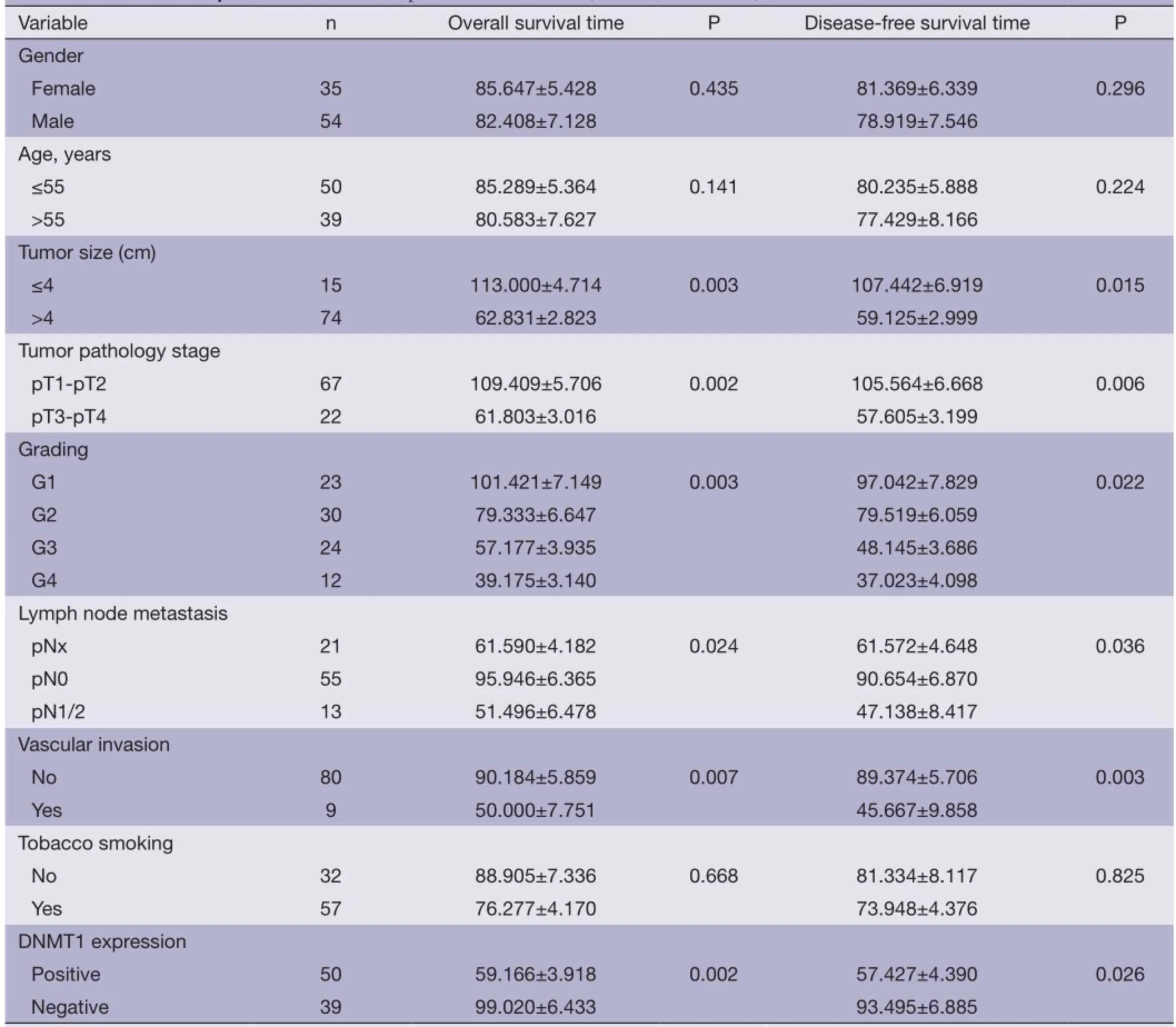
Table 4 Univariate analysis of OS and DFS in patients with ccRCC (months, mean ± SE)
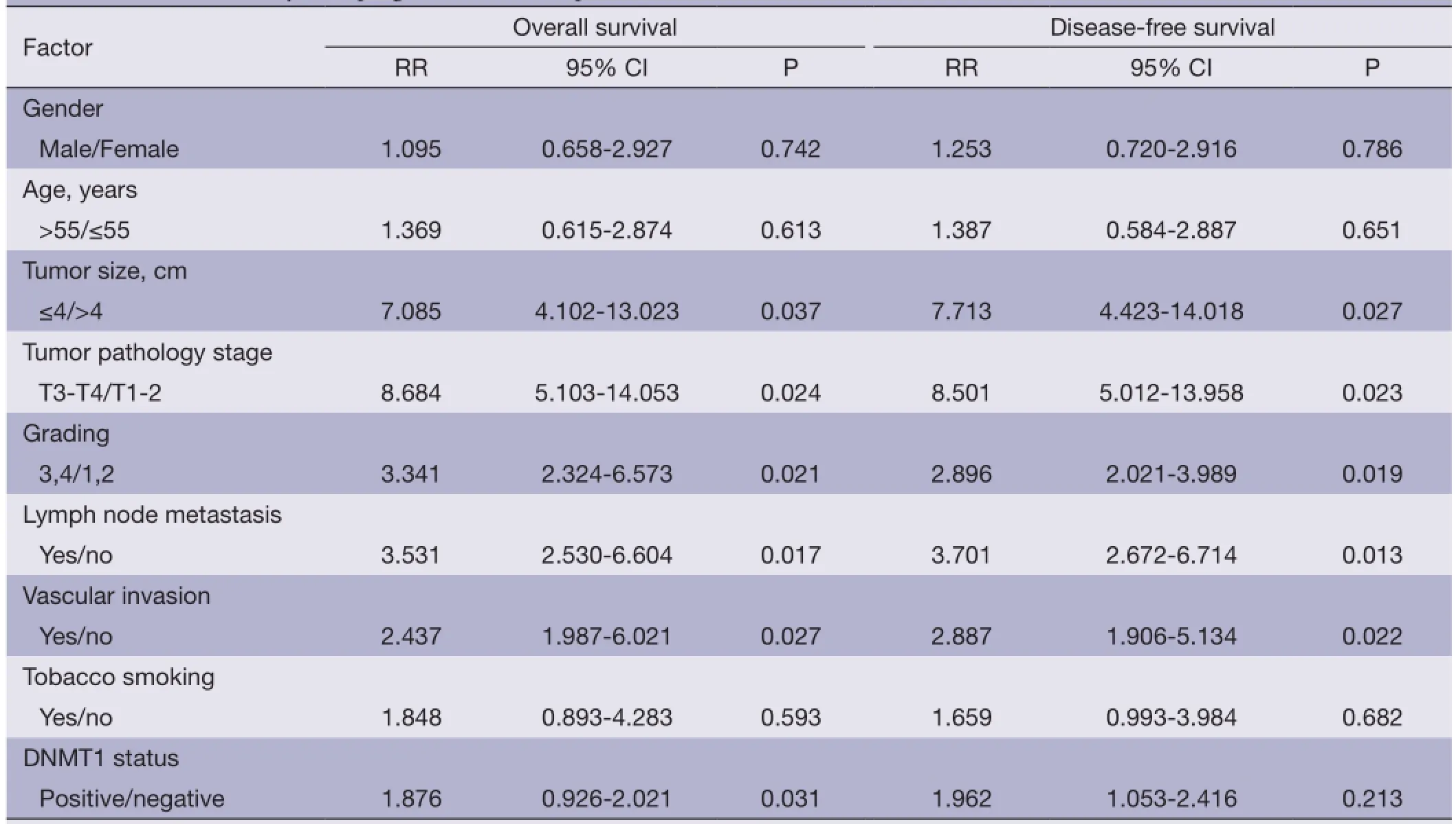
Table 5 Multivariate analysis of prognostic factors in patients with ccRCC
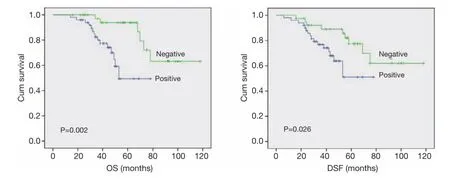
Figure 2 Univariate analyses of OS and DSF in patients with ccRCC using the Kaplan-Meier method according to DNMT1 expression. The log rank test was used to calculate P values. OS, overall survival; DFS, disease-free survival; ccRCC, clear cell renal cell carcinoma; DNMT1, DNA methyltransferase 1.
Discussion
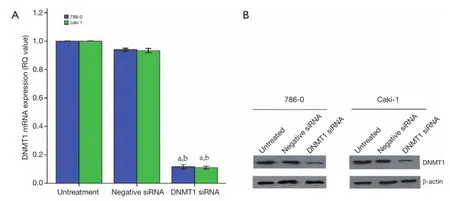
Figure 3 Konckdown of DNMT1 expressions in ccRCC cells. DNMT1 siRNA was transiently transfected into ccRCC cell lines and then subjected to qRT-PCR (A) and western blotting (B) analysis for DNMT1 expression. a, P<0.05 compared with the untreated cells. b, P<0.05 compared with the negative siRNA cells. The dates are shown as the mean ± SD of six independent experiments. DNMT1, DNA methyltransferase 1; ccRCC, clear cell renal cell carcinoma.

Figure 4 Effects of DNMT1 siRNA on cell viability (A), apoptosis (B), colony formation (C) and invading ability of invation (D). a, P<0.05 compared with the untreated cells. b, P<0.05 compared with the negative siRNA transfected cells. The data are shown as the mean ± SD of eight independent experiments. DNMT1, DNA methyltransferase 1.
In the present study, a comprehensive knowledge about the pathogenic characteristics of DNMT1 involved in ccRCC tumorigenesis was gathered and our data indicate that overexpression of DNMT1 plays a key role in ccRCC tumorigenesis and progression. Consistent with the previous studies in other types of cancer (10-12,23-26), wedemonstrated that DNMT1 expression was higher in the ccRCC than that in adjacent non-tumor tissues, suggesting that DNMT1 may be involved in the malignancy of ccRCC. This fnding is also agreeable to previous study reporting that DNMT1 expression was signifcantly higher in RCCs (21). In any events, due to the complex mechanisms responsible for regulation of DNMTs expressions and functions of DNMTs in carcinogenesis, the altered expression and effects of DNMTs should be further investigated in ccRCC although their aberrant expressions were found to be because of methylation of their gene promoters in different cancers, such as gliomas and embryonic tissues (27,28). In addition, microRNAs (miRs), which are noncoding RNAs, are also involved in regulation of DNMT expression (29-32).
Our current study further associated the relevance of DNMT1 protein expression with clinicopathological features from ccRCC patients. The data showed that DNMT1 protein expression was positively correlated with tumor size, clinical stage, histological grade suggesting that DNMT1 may be markers for tumor progression. In addition, tumor invasion is considered to be an important risk factor for metastatic ccRCC. In our study, the positive expression of DNMT1 protein was associated with a highly malignant phenotype and could be considered as a poor prognostic index for ccRCC. This observation was confrmed by the study in vitro, we found that knockdown of DNMT1 expression signifcantly inhibited the viability, colony formation and invading ability, induced apoptosis in ccRCC cells. Moreover, our multivariate analysis showed that patients with ccRCC exhibiting positive DNMT1 protein expression had a signifcantly poor over-all survival, suggesting that DNMT1 is a biologic predictor of poor prognosis of ccRCC patient. However, to date, we don’t know why these happened or the implication of these associations. Previous studies have demonstrated that hypermethylation of tumor suppress gene (TSG) promoters is crucial for cancer initiation and progression (33). Hypermethylation of some TSG promoters that affect the prognosis can partially explain the poor prognosis associated with DNMT overexpression (34-38).
Hypermethylation of tumor suppressor genes is caused by aberrant up-regulation of DNMT protein expression and enzyme activity. Thus, many researchers have targeted DNMT as a novel treatment strategy for tumors. Cytosine analogues capable of trapping DNMTs onto DNA, inhibition of DNMT mRNA expression through siRNA (39,40), antisense (40), irradiation (41) and mediation through the Hedgehog pathway (42) have been explored in previous studies. Recently in vitro studies have shown DNMTs inhibitor can induce apoptosis in RCC cells (43,44). In addition, another study suggest that DNMTs inhibitor could suppress RCC cell proliferation by inducing G2/M cell cycle arrest and strikingly increase the sensitivity of RCC to paclitaxel (45), leading to using DNMT inhibitors in clinical trials of renal cancers (46-48). Our data showed that knockdown of DNMT1 expression significantly inhibited cell viability, induced apoptosis, decreased colony formation and invading ability in ccRCC cells. These results were in agreement with previous studies in bladder cancer (13), pancreas cancer (49), colorectal cancer (50), ovarian cancer (51), cholangiocarcinoma (52), as well as in lung, esophageal cancer, and malignant pleural mesothelioma cells (53). Moreover, some studies have also investigated the synergistic knockdown of DNMT1 and DNMT3b and shown a synergistic effect in CP70 ovarian cancer cell line (51), QBC-939 cholangiocarcinoma cell line (52) and a colorectal cancer cell line (50). Thus, further studies are needed to confrm whether there are synergistic effects after combination of DNMT1 and DNMT3a or DNMT3b siRNA in ccRCC.
In summary, the present study demonstrated that DNMT1 was higher expressed in ccRCC than no-tumor tisuses, and the expression of DNMT1 was strongly associated with ccRCC tumor size, tumor pathology stage, histological grading, lymph node metastasis, vascular invasion, recurrence, and prognosis. Knockdown of DNMT1 expression significantly inhibited the viability and induced apoptosis of ccRCC cells, as well as inhibited the colony formation and invading ability. DNMT1 may thus serve as a potential prognostic marker and a novel therapeutic target for ccRCC patients. Nevertheless, further studies with large samples are warranted to confrm the present fndings.
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (No. 30873097). We thank Shanye Yin and Ja Ya for their excellent language editing.
Disclosure: The authors declare no confict of interest.
1. Miyamoto H, Miller JS, Fajardo DA, et al. Non-invasive papillary urothelial neoplasms: the 2004 WHO/ISUP classifcation. Pathol Int 2010;60:1-8.
2. Karakiewicz PI, Briganti A, Chun FK, et al. Multi-institutional validation of a new renal cancer-specifc survival nomogram. J Clin Oncol 2007;25:1316-22.
3. Matsuura K, Nakada C, Mashio M, et al. Downregulation of SAV1 plays a role in pathogenesis of high-grade clear cell renal cell carcinoma. BMC Cancer 2011;11:523.
4. Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 2003;349:2042-54.
5. Robertson KD. DNA methylation and chromatin -unraveling the tangled web. Oncogene 2002;21:5361-79.
6. Hermann A, Gowher H, Jeltsch A. Biochemistry and biology of mammalian DNA methyltransferases. Cell Mol Life Sci 2004;61:2571-87.
7. Robert MF, Morin S, Beaulieu N, et al. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet 2003;33:61-65.
8. Hino R, Uozaki H, Murakami N, et al. Activation of DNA Methyltransferase 1 by EBV Latent Membrane Protein 2A Leads to Promoter Hypermethylation of PTEN Gene in Gastric Carcinoma. Cancer Res 2009;69:2766.
9. Girault I, Tozlu S, Lidereau R, et al. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Cancer Res 2003;9:4415-22.
10. Saito Y, Kanai Y, Nakagawa T, et al. Increased protein expression of DNA methyltransferase (DNMT) 1 is signifcantly correlated with the malignant potential and poor prognosis of human hepatocellular carcinomas. Int J Cancer 2003;105:527-32.
11. Lin RK, Hsu HS, Chang JW, et al. Alteration of DNA methyltransferases contributes to 5'CpG methylation and poor prognosis in lung cancer. Lung Cancer 2007;55:205-13.
12. Kanai Y, Ushijima S, Kondo Y, et al. DNA methyltransferase expression and DNA methylation of CPG islands and peri-centromeric satellite regions in human colorectal and stomach cancers. Int J Cancer 2001;91:205-212.
13. Wu CT, Wu CF, Lu CH, et al. Expression and function role of DNA methyltransferase 1 in human bladder cancer. Cancer 2011;117:5221-33.
14. Gravina GL, Ranieri G, Muzi P, et al. Increased levels of DNA methyltransferases are associated with the tumorigenic capacity of prostate cancer cells. Oncol Rep 2013;29:1189-95.
15. Tessema M, Länger F, Dingemann J, et al. Aberrant methylation and impaired expression of the p15(INK4b) cell cycle regulatory gene in chronic myelomonocytic leukemia (CMML). Leukemia 2003;17:910-8.
16. Sun L, Hui AM, Kanai Y, et al. Increased DNA methyltransferase expression is associated with an early stage of human hepatocarcinogenesis. Jpn J Cancer Res 1997;88:1165-70.
17. Fujii S, Katake Y, Tanaka H. Increased expression of DNA methyltransferase-1 in non-neoplastic epithelium helps predict colorectal neoplasia risk in ulcerative colitis. Digestion 2010;82:179-86.
18. Amara K, Ziadi S, Hachana M, et al. DNA methyltransferase DNMT3b protein overexpression as a prognostic factor in patients with diffuse large B-cell lymphomas. Cancer Sci 2010;101:1722-30.
19. Sawada M, Kanai Y, Arai E, et al. Increased expression of DNA methyltransferase 1 (DNMT1) protein in uterine cervix squamous cell carcinoma and its precursor lesion. Cancer Lett 2007;251:211-9.
20. Peng DF, Kanai Y, Sawada M, et al. Increased DNA methyltransferase 1 (DNMT1) protein expression in precancerous conditions and ductal carcinomas of the pancreas. Cancer Sci 2005;96:403-8.
21. Arai E, Kanai Y, Ushijima S, et al. Regional DNA hypermethylation and DNA methyltransferase (DNMT) 1 protein overexpression in both renal tumors and corresponding nontumorous renal tissues. Int J Cancer 2006;119:288-96.
22. Eble JN, Sauter G, Epstein JI, et al. eds. Pathology and Genetics of Tumours of the Urinary System and Male Genital Organs. Lyon: IARC Press, 2004.
23. Yang J, Wei X, Wu Q, et al. Clinical signifcance of the expression of DNA methyltransferase proteins in gastric cancer. Mol Med Rep 2011;4:1139-43.
24. Qu Y, Mu G, Wu Y, et al. Overexpression of DNA methyltransferases 1, 3a, and 3b signifcantly correlates with retinoblastoma tumorigenesis. Am J Clin Pathol 2010;134:826-34.
25. Xing J, Stewart DJ, Gu J, et al. Expression of methylationrelated genes is associated with overall survival in patients with non-small cell lung cancer. Br J Cancer 2008;98:1716-22.
26. Park HJ, Yu E, Shim YH. DNA methyltransferase expression and DNA hypermethylation in human hepatocellular carcinoma. Cancer Lett 2006;233:271-8.
27. Rajendran G, Shanmuganandam K, Bendre A, et al. Epigenetic regulation of DNA methyltransferases: DNMT1 and DNMT3B in gliomas. Journal of Neuro-Oncology 2011;104:483-94.
28. Novakovic B, Wong NC, Sibson M, et al. DNA methylation-mediated down-regulation of DNA methyltransferase-1 (DNMT1) is coincident with, but not essential for, global hypomethylation in human placenta. J Biol Chem 2010;285:9583-93.
29. Fabbri M, Garzon R, Cimmino A, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc NatlAcad Sci U S A 2007;104:15805-10.
30. Huang J, Wang Y, Guo Y, et al. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology 2010;52:60-70.
31. Duursma AM, Kedde M, Schrier M, et al. miR-148 targets human DNMT3b protein coding region. RNA 2008;14:872-7.
32. Chen KC, Wang YS, Hu CY, et al. OxLDL up-regulates microRNA-29b, leading to epigenetic modifcations of MMP-2/MMP-9 genes: a novel mechanism for cardiovascular diseases. FASEB J 2011;25:1718-28.
33. Ron-Bigger S, Bar-Nur O, Isaac S, et al. Aberrant epigenetic silencing of tumor suppressor genes is reversed by direct reprogramming. Stem Cells 2010;28:1349-54.
34. Wu D, Xiong L, Wu S, et al. TFPI-2 methylation predicts poor prognosis in non-small cell lung cancer. Lung Cancer 2012;76:106-11.
35. Xu L, Li X, Chu ES, et al. Epigenetic inactivation of BCL6B, a novel functional tumour suppressor for gastric cancer, is associated with poor survival. Gut 2012;61:977-85.
36. Zhou W, Jiang Z, Liu N, et al. Down-regulation of CXCL12 mRNA expression by promoter hypermethylation and its association with metastatic progression in human breast carcinomas. J Cancer Res Clin Oncol 2009;135:91-102.
37. Chen HY, Zhu BH, Zhang CH, et al. High CpG island methylator phenotype is associated with lymph node metastasis and prognosis in gastric cancer. Cancer Sci 2012;103:73-9.
38. Zhang Q, Chen L, Helfand BT, et al. TGF-β regulates DNA methyltransferase expression in prostate cancer, correlates with aggressive capabilities, and predicts disease recurrence. PLoS One 2011;6:e25168.
39. Xu M, Gao J, Du YQ, et al. Reduction of pancreatic cancer cell viability and induction of apoptosis mediated by siRNA targeting DNMT1 through suppression of total DNA methyltransferase activity. Mol Med Rep 2010;3:699-704.
40. Przybylski M, Kozłowska A, Pietkiewicz PP, et al. Increased CXCR4 expression in AsPC1 pancreatic carcinoma cells with RNA interference-mediated knockdown of DNMT1 and DNMT3B. Biomed Pharmacother 2010;64:254-8.
41. Ma JX, Jin ZD, Si PR, et al. Continuous and low-energy 125I seed irradiation changes DNA methyltransferases expression patterns and inhibits pancreatic cancer tumor growth. J Exp Clin Cancer Res 2011;30:35.
42. He S, Wang F, Yang L, et al. Expression of DNMT1 and DNMT3a are regulated by GLI1 in human pancreatic cancer. PLoS One 2011;6:e27684.
43. Konac E, Varol N, Yilmaz A, et al. DNA methyltransferase inhibitor-mediated apoptosis in the Wnt/β-catenin signal pathway in a renal cell carcinoma cell line. Exp Biol Med (Maywood) 2013;238:1009-16.
44. Gu B, Ding Q, Xia G, et al. EGCG inhibits growth and induces apoptosis in renal cell carcinoma through TFPI-2 overexpression. Oncol Rep 2009;21:635-40.
45. Shang D, Ito N, Kamoto T, et al. Demethylating agent 5-aza-2'-deoxycytidine enhances susceptibility of renal cell carcinoma to paclitaxel. Urology 2007;69:1007-12.
46. Stewart DJ, Donehower RC, Eisenhauer EA, et al. A phase I pharmacokinetic and pharmacodynamic study of the DNA methyltransferase 1 inhibitor MG98 administered twice weekly. Ann Oncol 2003;14:766-74.
47. Winquist E, Knox J, Ayoub JP, et al. Phase II trial of DNA methyltransferase 1 inhibition with the antisense oligonucleotide MG98 in patients with metastatic renal carcinoma: a National Cancer Institute of Canada Clinical Trials Group investigational new drug study. Invest New Drugs 2006;24:159-67.
48. Amato RJ, Stephenson J, Hotte S, et al. MG98, a secondgeneration DNMT1 inhibitor, in the treatment of advanced renal cell carcinoma. Cancer Invest 2012;30:415-21.
49. Gao J, Wang L, Xu J, et al. Aberrant DNA methyltransferase expression in pancreatic ductal adenocarcinoma development and progression. J Exp Clin Cancer Res 2013;32:86.
50. Rhee I, Bachman KE, Park BH, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 2002;416:552-6.
51. Leu YW, Rahmatpanah F, Shi H, et al. Double RNA interference of DNMT3b and DNMT1 enhances DNA demethylation and gene reactivation. Cancer Res 2003;63:6110-5.
52. Zuo S, Luo J, Liu M, et al. Suppressing effects of downregulating DNMT1 and DNMT3b expression on the growth of human cholangiocarcinoma cell line. J Huazhong Univ Sci Technolog Med Sci 2008;28:276-80.
53. Kassis ES, Zhao M, Hong JA, et al. Depletion of DNA methyltransferase 1 and/or DNA methyltransferase 3b mediates growth arrest and apoptosis in lung and esophageal cancer and malignant pleural mesothelioma cells. J Thorac Cardiovasc Surg 2006;131:298-306.
Cite this article as:Li M, Wang Y, Song Y, Bu R, Yin B, Fei X, Guo Q, Wu B. Aberrant DNA methyltransferase 1 expression in clear cell renal cell carcinoma development and progression. Chin J Cancer Res 2014;26(4):371-381. doi: 10.3978/ j.issn.1000-9604.2014.08.03
10.3978/j.issn.1000-9604.2014.08.03
Submitted Apr 24, 2014. Accepted for publication Jul 30, 2014.
杂志排行
Chinese Journal of Cancer Research的其它文章
- In vitro effect of iASPP on cell growth of oral tongue squamous cell carcinoma
- Long-term survival outcomes of video-assisted thoracic surgery for patients with non-small cell lung cancer
- Embolization of symptomatic renal angiomyolipoma with a mixture of lipiodol and PVA, a mid-term result
- Decline of serum CA724 as a probable predictive factor for tumor response during chemotherapy of advanced gastric carcinoma
- TPX2 knockdown suppressed hepatocellular carcinoma cell invasion via inactivating AKT signaling and inhibiting MMP2 and MMP9 expression
- Safety and feasibility of video-assisted thoracoscopic surgery for stage IIIA lung cancer
