Morphological,cytological and molecular analyses of a synthetic hexaploid derived from an interspecific hybrid between Gossypium hirsutum and Gossypium anomalum
2014-03-13XiaZhangCaijiaoZhaiLinhiHeQiGuoXiangguiZhangPengXuHongmeiSuYuanyongGongWanhaoNiXinlianShen
Xia Zhang,Caijiao Zhai,Linhi He,Qi Guo,Xianggui Zhang,Peng Xu,Hongmei Su,Yuanyong Gong,Wanhao Ni,Xinlian Shen,*
aKey Laboratory of Cotton and Rapeseed,Ministry of Agriculture,Nanjing,China
bThe Institute of Industrial Crops,Jiangsu Academy of Agricultural Sciences,Nanjing 210014,China
cJiangsu Yanjiang Institute of Agricultural Sciences,Nantong 226541,China
1.Introduction
The genus Gossypium encompasses 50 species (45 diploids and five allopolyploids),which are distributed throughout most tropical and subtropical regions of the world[1].Of the four cultivated species,Gossypium hirsutum L.(2n = 4x = 52,A1D1) is responsible for approximately 90% of the total cotton production worldwide.The other three principal cultivated species are the African diploid Gossypium herbaceum L.(2n = 2x = 26,A2),the Asian and Indian diploid Gossypium arboreum L.(2n = 2x = 26,A1),and the New World tetraploid Gossypium barbadense L.(2n = 4x = 52,A2D2).Diploid Gossypium species fall into eight cytological groups,designated A–G and K based on their chromosomal pairing relationships and geographical distribution [2,3].Wild species of cotton represent a significant genetic repository for potential exploitation by cotton breeders,who have long recognized the beneficial effects of exotic genes [4].The introduction of alien genetic variation into upland cotton from the chromosomes of wild species is a valuable and proven technique for cotton improvement.The most successful examples of the use of wild species during the history of cotton breeding history include Gossypium harknessii as a source of cytoplasmic male sterility [5] and Gossypium thurberi as a source of fiber quality [6,7].More recently,other important traits,such as nematode resistance and the low-and high-gossypol plant traits,were successfully introduced from diploid species into upland cotton using various strategies [8,9].Despite these successes,most of the genetic variation in wild Gossypium species remains to be exploited.
G.anomalum(2n = 2x = 26,B1)is a wild species belonging to the B1genome group.G.anomalum grows in Southwest Africa and along the southern fringes of the Sahara,almost from the Atlantic to the Red Sea [1].As a member of subsection Anomala Todaro,G.anomalum possesses several desirable characters such as extremely fine fibers,good strength,low fiber weight,resistance to insect pests,immunity to the diseases black arm and bacterial blight and tolerance to water deficit,as this species is endemic to relatively dry areas [10].Some efforts have been made to introduce desirable characters from G.anomalum to cultivated cotton[11,12].
G.anomalum represents an inestimable source of genes that can potentially be transferred to the cultivated cotton gene pool.However,the genomic differences between tetraploid cultivated cotton (A1A1D1D1) and the diploid G.anomalum (B1B1)represent serious interspecific reproductive barriers,which limit gene transfer between the species.In a previous study,we obtained triploid hybrids with the genome composition A1D1Blby crossing G.hirsutum (A1A1D1D1) with G.anomalum (B1B1) [11].Hybrid seedling plants were then treated with 0.15% colchicine and a putative fertile hexaploid (A1A1D1D1B1B1) was obtained.This putative hexaploid produced flowers and set bolls normally.The objectives of this study were: (1) to confirm the hexaploid nature of the plants using morphological,cytological and molecular methods;(2)to compare EST-SSR transferability from other species to G.anomalum;and(3)to obtain a set of informative G.anomalum-specific SSR markers to monitor G.anomalum-specific chromosome segments.
2.Materials and methods
2.1.Experimental materials
Seedling plants of triploid hybrids from the cross between G.hirsutum (A1A1D1D1) var.86-1 and G.anomalum (B1B1) were treated with 0.15%colchicine[11].A putative fertile hexaploid(A1A1D1D1B1B1) was selfed and the resulting hexaploid seeds were stored in a-20 °C freezer.In 2009,all experimental materials,including the putative hexaploid,G.anomalum and G.hirsutum var.86-1,were planted in pots at the Lishui Experiment Station,Jiangsu Academy of Agricultural Science,and maintained in a greenhouse through the winter.Seeds were harvested in the greenhouse in the spring of 2010 and used in mitotic chromosome preparation.
2.2.Cytological analysis
To check the ploidy level of the putative hexaploid hybrid,mitotic chromosome preparations were carried out using root tips.Roots were excised from germinated seedlings on MS medium when they were approximately 3 cm long,pretreated with 0.025% (v/v) cycloheximide at room temperature for 2 h to accumulate metaphase cells,and fixed in Carnoy's solution(ethanol:acetic acid = 3:1,v/v).The root tips were macerated in 2% cellulose and 0.5% pectinase at 37 °C for 40 min and squashed on slides in 60%acetic acid.All slides were stored at-70 °C overnight.The slides were then stained in 6-diamidino-2-phenylindole (Roche Diagnostics) for 3 min at room temperature and examined under an Olympus BX51 fluorescence microscope.Between 20 and 30 cells in each of the putative hexaploid hybrid plants were examined for chromosome number.
2.3.SSR analysis
Genomic DNA from four putative hexaploid plants and the parental accessions were extracted as described by Paterson et al.[13].A total of 707 SSR primer pairs covering the whole cotton genome were selected based on the cotton reference map [14] and marker chromosome location information [15].The sequences of these primers are available from the Cotton Marker Database (CMD) (http://www.cottonmarker.org/).SSR analysis was conducted according to Zhang et al.[16].
3.Results
3.1.Morphological observations
Most of morphological characteristics of the putative hexaploid plants were intermediate between G.hirsutum and G.anomalum (Fig.1); for example the shapes and sizes of leaves,bolls and bracts of hexaploid plants.The hexaploid plants had large petal spots and intense hairiness inherited from G.anomalum.They exhibited prolific growth,and developed many bolls,with 5–13 seeds in every capsule.However,when they were used as male parents in backcrosses to G.hirsutum,seeds were rarely obtained.

Fig.1-Morphological characteristics of G.anomalum,G.hirsutum,and the(G.hirsutum × G.anomalum)2 hexaploid.a:G.anomalum;b:G.hirsutum var.86-1;c: (G.hirsutum × G.anomalum)2 hexaploid.I:adult plants;II:fully opened flowers;III: petals,pistil and bracts;IV: mature leaves;V:fuzzy seeds.
3.2.Mitotic chromosome analysis
Mitotic metaphase counts revealed the presence of 78 chromosomes in all four plants of the (G.hirsutum × G.anomalum)2hexaploid (Fig.2) confirming the amphiploid status of the material because it is in agreement with the number of chromosomes expected for a synthetic hexaploid (2n = 6x =78,A1A1D1D1B1B1) resulting from a cross of G.hirsutum (2n =4x = 52,A1A1D1D1)and G.anomalum(2n = 2x = 26,B1B1).
3.3.Analysis SSR band patterns in G.hirsutum,G.anomalum,and the hexaploid hybrid
A total of 707 SSR primer pairs covering the cotton genome were selected to amplify the two parents and four hexaploid hybrid plants.Among them,94 were developed from G.arboreum EST sequences,378 from Gossypium raimondii EST sequences and 235 from G.hirsutum EST sequences [14,15].All 707 primer pairs yielded microsatellite products in G.hirsutum var.86-1 and in the hexaploid hybrid plants; 683 produced polymorphic bands between G.hirsutum var.86-1 and G.anomalum,10 were monomorphic between the two parents and 14 failed to produce a PCR product in G.anomalum.A high level of polymorphism (96.6%) was observed between G.hirsutum and G.anomalum,confirming that these two species are genetically distant.Among the 683 polymorphic primer pairs,674(98.68%)had an additive banding pattern in the hexaploid,indicating the hybrid status of the hexaploidata genome-wide level; 9 SSR primer pairs failed to amplify G.anomalum-specific bands in the hexaploid plants.
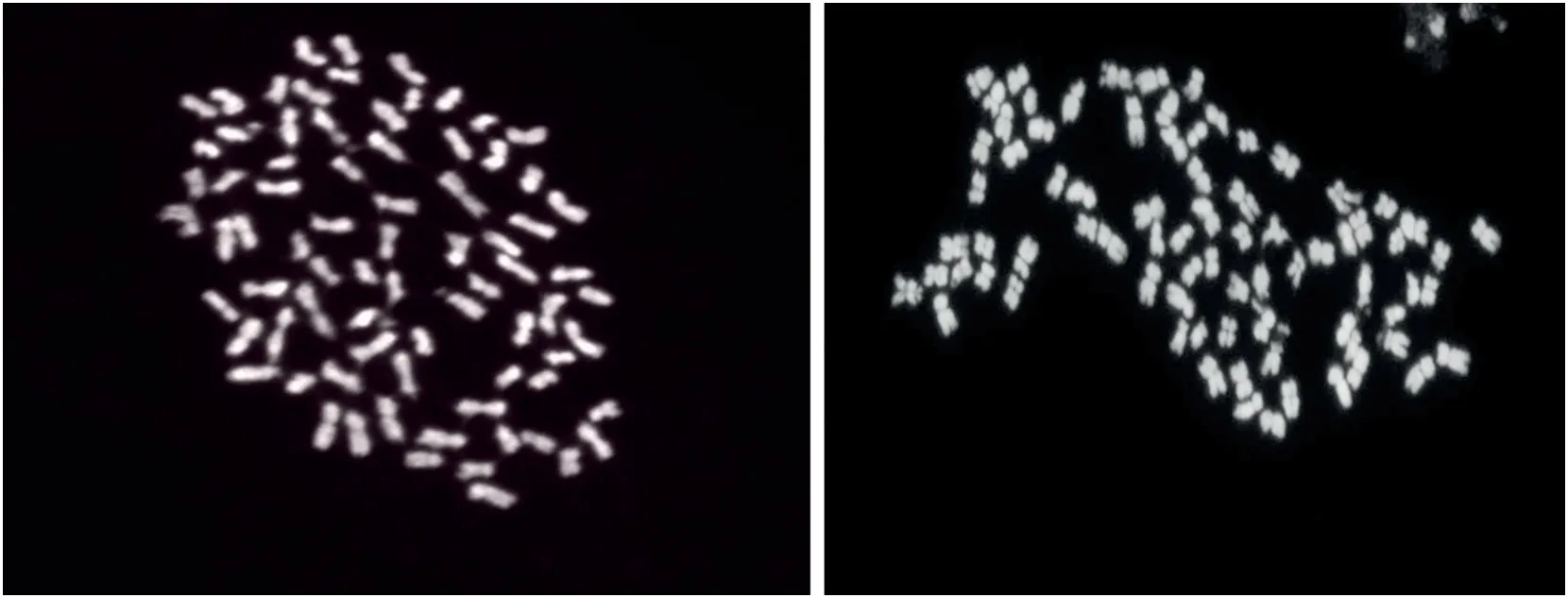
Fig.2-Mitotic metaphase in hexaploid plants,showing 78 chromosomes.
Four hundred and twelve markers (58.3%) yielded easily distinguishable microsatellite products.The number of bands per SSR marker in G.hirsutum and G.anomalum was scored based on dominant scoring of the SSR bands,characterized by the presence or absence of a particular band.In G.hirsutum,the 412 markers produced 1499 bands,averaging 3.87 bands per marker,whereas in G.anomalum,they produced 815 bands,averaging 2.2 bands per marker.There were 457 common bands between G.hirsutum and G.anomalum,averaging 1.22 common bands per marker.A-genome-derived markers produced more common bands(1.35 bands per markers)than D-genome-derived markers(1.18 bands per marker)(Table 1).
The polymorphisms of SSR loci between G.hirsutum and G.anomalum appeared as five types of basic banding patterns in the hexaploid hybrid.Of the 683 EST-SSRs that produced polymorphic amplifications,333 (47.1%) displayed pattern A,where the polymorphic bands in the hexaploid hybrid were shared by both parents (codominant loci).A-genome-derived markers produced more codominant loci than D-genomederived markers (Table 2).A total of 334 (47.24%) markers displayed pattern B,in which the polymorphic bands in the hexaploid hybrid were from G.hirsutum (dominant in G.hirsutum),whereas 16 markers displayed pattern C,in which the polymorphic bands in the hexaploid hybrid were from G.anomalum (dominant in G.anomalum) (Table 2).There were two other extreme instances of band pattern,including one instance in which G.anomalum produced no bands and one in which G.anomalum-specific bands were not amplified in the hexaploid hybrid plants.Among the 14 primer pairs that failed to produce a PCR product in G.anomalum,two were A-genome-derived,10 were D-genome-derived,and two were AD-genome-derived,indicating that A-genome-derived SSR markers have a higher level of transferability than D-genomederived SSR markers in G.anomalum.In addition,there were nine SSR primer pairs(NAU2139,NAU2169,NAU2182,NAU2954,NAU3119,NAU3317,NAU3480,NAU3489,and NAU5152) that produced no G.anomalum-specific bands in the hexaploid plants.As G.hirsutum will be used as the recurrent parent in backcrossing programs,those dominant loci in G.hirsutum cannot be used to monitor introgression of G.anomalum-specific segments during backcrossing.Therefore,a total of 349 informative SSR markers (333 codominant loci and 16 dominant loci in G.anomalum)can be used in future backcrossing programs.
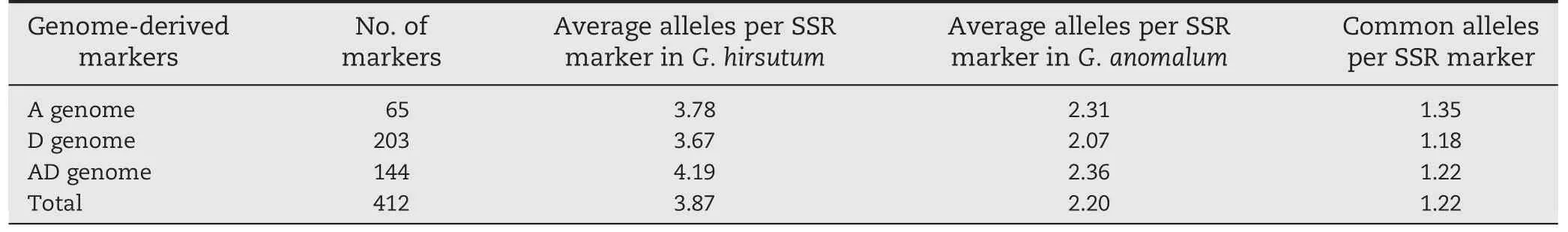
Table 1-Comparison of SSR alleles between G.hirsutum and G.anomalum.
4.Discussion
EST-SSR markers derived from transcribed regions of DNA were shown to produce high rates of transferability in cotton species [17] and other related plant groups [18,19].In this study,a total of 707 SSR markers developed from G.arboreum(A genome),G.raimondii(D genome),and G.hirsutum(AD genome)were chosen to amplify DNA from G.hirsutum,G.anomalum,and the synthetic.All 707 primer pairs yielded microsatellite products in G.hirsutum and the synthetic,but 14 failed to produce a band in G.anomalum.However the transferability rate from the three species to G.anomalum was very high(98.0%).D-genome-derived SSR markers showed slightly lower rates of transferability than A-and AD-genome species-derived markers.Although all selected SSR markers expressed high levels of transferability and polymorphism,almost half of the markers (47.24%) were dominant in G.hirsutum.Since G.hirsutum will be used as the recurrent parent in future backcrossing programs,those dominant markers in G.hirsutum cannot be used to monitor the introgression of G.anomalum-specific segments in backcross populations.However,the A-genome-derived markers produced more codominant loci (56.38%) than the D-genome-derived markers(42.59%),indicating that the A-genome-derived markers were more powerful for distinguishing genomic differences between G.hirsutum and G.anomalum.In addition,the A-genomederived SSR markers have a higher level of transferability between G.hirsutum and G.anomalum than the D-genomederived SSR markers.These phenomena suggest that SSR markers developed from close relatives of the wild species are more applicable to the analysis of the transfer of chromosome segments from the wild species to cultivated cotton than other types of markers.
Although hybrids are expected to have additive banding profiles of the two parents,previous work has demonstrated that allopolyploid speciation in plants may be associated with non-Mendelian genomic changes in the early generations following polyploid synthesis in crops such as wheat [20,21]and rapeseed[22].However,there is no evidence for structural genomic changes or de novo DNA methylation modifications in newly synthesized allopolyploid cotton[23].In this study,9 SSR primer pairs failed to amplify G.anomalum-specific bands in hexaploid plants.Possible explanations for this include chromosome loss,heterozygosity at some loci in a parental plant,chromosome rearrangement,and sequence discrepancy between the different species.Loss of chromosomes was not the causative factor because the synthetic plants had the expected chromosome number (2n = 78).The presence of heterozygous loci in a parental plant was also unlikely since there was no evidence of variation among the tested parental plants.We also ruled out the possibility of chromosome rearrangement since one SSR marker(NAU2954)that failed to amplify G.anomalum-specific bands in the synthetic is closely linked to two markers(NAU2580 and NAU5373)on the Guo et al.[14] genetic map that did amplify as expected in the synthetic.Sequence discrepancy between the different species may be a likely reason for failure of expression of some SSR bands in the hexaploid hybrid.The SSR primers used were based on sequences of G.arboreum (A1genome),G.raimondii(D5genome) and G.hirsutum (A1D1genome).Compared to these species,many base substitutions may have occurred in the flanking sequences adjacent to the SSR loci in G.anomalum(B1genome); some of these substitutions may have been in the marker binding sites thus causing a preferential primer binding to genomic DNA from G.hirsutum.As a result,the specific SSR bands of G.anomalum would be undetectable in the hybrid plants.This explanation was confirmed by the observation that amplification of specific SSR bands of G.anomalum also failed when a DNA pool from G.hirsutum and G.anomalum was used instead of synthetic DNA as the template(Fig.3).

Table 2-Comparison of amplification patterns produced with different genome-derived SSR primers.
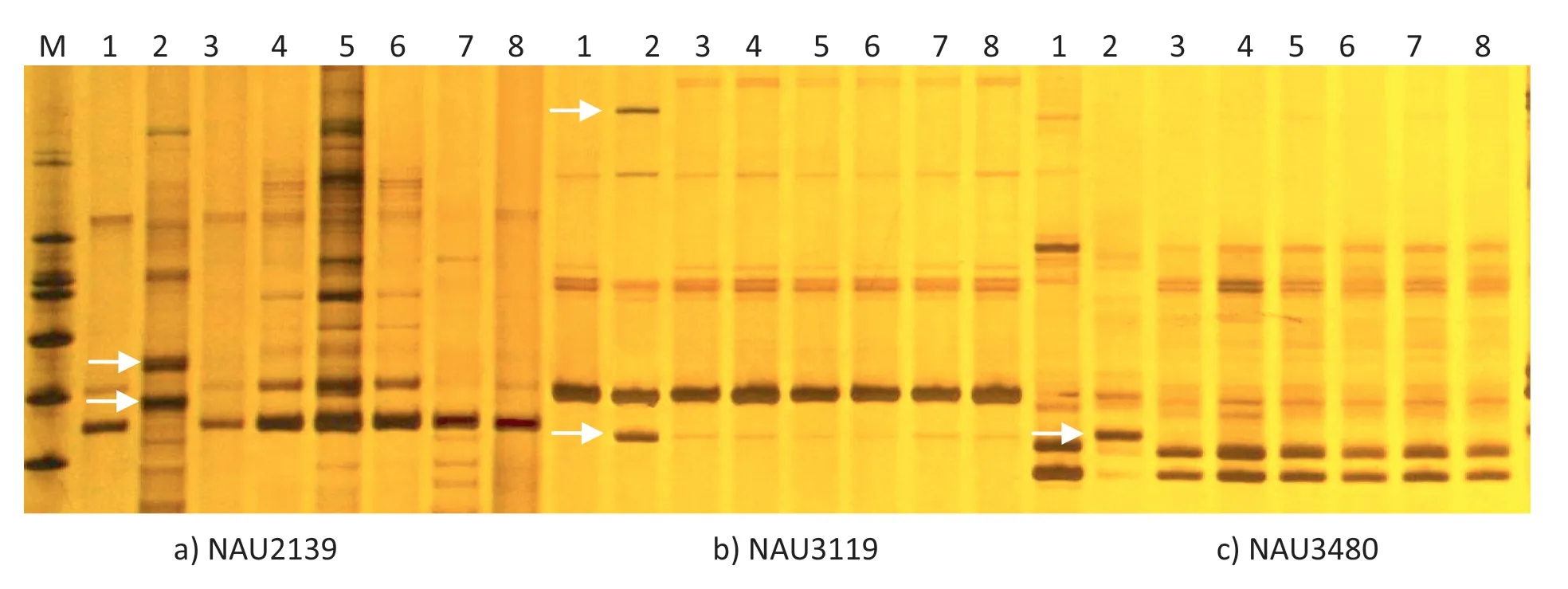
Fig.3-Amplification patterns of G.anomalum-specific bands for three SSR markers showing with amplification failure in the synthetic.M:marker;1:G.hirsutum var.86-1;2:G.anomalum;3-6:(G.hirsutum × G.anomalum)2 synthetic;7-8:DNA pool of G.hirsutum and G.anomalum.Arrows indicate bands that failed to amplify in the synthetic and in a DNA mixture from the parents.
The present results demonstrate the hybridity and doubled status of (G.anomalum × G.hirsutum) using morphological,cytological and molecular marker methods.These materials can be used as bridges for the transfer of useful agronomic traits from the wild species to cultivated varieties.The 349 informative SSR markers generated for the interspecific hybrids can be used to track the flow of genetic material from G.anomalum during backcrossing to G.hirsutum.
This work was supported by the National Natural Science Foundation of China(31171595)and the Independent Innovation Funds for Agricultural Technology of Jiangsu Province,China[CX(12)5039].
[1] P.A.Fryxell,A revised taxonomic interpretation of Gossypium L.(Malvaceae),Rheedea 2(1992) 108–165.
[2] J.F.Wendel,New World tetraploid cottons contain old world cytoplasm,Proc.Natl.Acad.Sci.U.S.A.86(1989) 4132–4136.
[3] A.E.Percival,J.F.Wendel,J.M.Stewart,Taxonomy and germplasm resources,in:C.W.Smith,J.Cothren (Eds.),Cotton: Origin,History,Technology,and Production,John Wiley &Sons,Inc.,NY,USA,1999,pp.33–63.
[4] J.F.Wendel,C.L.Brubaker,T.Seelanan,Chapter 1: the origin and evolution of Gossypium,in:J.M.Stewart,D.Oosterhuis,J.J.Heitholt,J.R.Mauney (Eds.),Physiology of Cotton,Springer,Netherlands,2010,pp.1–18.
[5] V.G.Meyer,Male sterility from Gossypium harknessii,J.Hered.66(1975) 23–27.
[6] T.W.Culp,D.C.Harrell,Breeding methods for improving yield and fiber quality of upland cotton (Gossypium hirsutum L.),Crop Sci.13 (1973) 686–689.
[7] T.W.Culp,D.C.Harrell,T.Kerr,Some genetic implications in the transfer of high fiber strength genes to upland cotton,Crop Sci.19 (1979) 481–484.
[8] E.J.Sacks,A.F.Robinson,Introgression of resistance to reniform nematode (Rotylenchulus reniformis) into upland cotton (Gossypium hirsutum) from Gossypium arboretum and a G.hirsutum/Gossypium aridum bridging line,Field Crops Res.112 (2009) 1–6.
[9] H.Benbouza,J.M.Lacape,J.M.Jacquemin,B.Courtois,F.B.H.Diouf,D.Sarr,N.Konan,J.P.Baudoin,G.Mergeai,Introgression of the low-gossypol seed &high-gossypol plant trait in upland cotton: analysis of [(Gossypium hirsutum × G.raimondii)2× G.sturtianum] trispecific hybrid and selected derivatives using mapped SSRs,Mol.Breed.25(2010)273–286.
[10] S.N.Ganesh,P.C.Vivek,S.M.Subhash,S.J.Ashok,Interspecific hybridization in Gossypium L.: characterization of progenies with different ploidy-confirmed multigenomic backgrounds,Plant Breed.132 (2013) 211–216.
[11] S.Y.Qian,J.Q.Huang,Y.J.Peng,B.L.Zhou,M.C.Ying,D.Z.Shen,G.L.Liu,T.X.Hu,Y.J.Xu,L.M.Gu,W.C.NI,S.Cheng,Studies on the hybrid of Gossypium hirsutum L.and G.anomalum and application in breeding,Sci.Agric.Sin.25(1992) 44–51 (in Chinese with English abstract).
[12] S.S.Mehetre,Wild Gossypium anomalum:a unique source of fibre fineness and strength-overview and achievements of pre-breeding efforts,Curr.Sci.99(2010) 58–71.
[13] A.H.Paterson,E.S.Lander,J.D.Hewitt,S.Peterson,S.E.Lincoln,S.D.Tanksley,A rapid method for extraction of cotton(Gossypium spp.)genomic DNA suitable for RFLP or PCR analysis,Plant Mol.Biol.Report.11(1993) 122–127.
[14] W.Z.Guo,C.P.Cai,C.B.Wang,Z.G.Han,X.L.Song,K.Wang,X.W.Niu,C.Wang,K.Y.Lu,B.Shi,T.Z.Zhang,A microsatellite-based,gene-rich linkage map reveals genome structure,function and evolution in Gossypium,Genetics 176(2007) 527–541.
[15] J.Xiao,K.Wu,D.D.Fang,D.M.Stelly,J.Yu,R.G.Cantrell,New SSR markers for use in cotton(Gossypium spp.)improvement,J.Cotton Sci.13(2009) 75–157.
[16] J.Zhang,Y.T.Wu,W.Z.Guo,T.Z.Zhang,Fast screening of SSR markers in cotton with PAGE/silver staining,Cotton Sci.12(2000) 267–269 (in Chinese with English abstract).
[17] W.Z.Guo,W.Wang,B.L.Zhou,T.Z.Zhang,Cross-species transferability of G.arboreum-derived EST-SSRs in the diploid species of Gossypium,Theor.Appl.Genet.112 (2006)1573–1581.
[18] R.Bandopadhyay,S.Sharma,S.Rustgi,R.Singh,A.Kumar,H.S.Balyan,P.K.Gupta,DNA polymorphism among 18 species of Triticum–Aegilops complex using wheat EST-SSRs,Plant Sci.166 (2004) 349–356.
[19] M.C.Saha,M.A.R.Mian,I.Eujayl,J.C.Zwonitzer,L.J.Wang,G.D.Mayl,Tall fescue EST-SSR markers with transferability across several grass species,Theor.Appl.Genet.109 (2004)783–791.
[20] M.Feldman,B.Liu,G.Segal,S.Abbo,A.A.Levy,J.M.Vega,Rapid elimination of low-copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes,Genetics 147(1997)1381–1387.
[21] K.Kashkush,M.Feldman,A.A.Levy,Gene loss,silencing and activation in a newly synthesized wheat allotetraploid,Genetics 160 (2002) 1651–1659.
[22] K.Song,P.Lu,K.Tang,T.C.Osborn,Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution,Proc.Natl.Acad.Sci.U.S.A.92(1995)7719–7723.
[23] B.Liu,C.L.Brubaker,G.Mergeai,R.C.Cronn,J.F.Wendel,Polyploid formation in cotton is not accompanied by rapid genomic changes,Genome 44(2001) 321–330.
杂志排行
The Crop Journal的其它文章
- An expedited method for isolation of DNA for PCR from Magnaporthe oryzae stored on filter paper
- Genetic dissection of tetraploid cotton resistant to Verticillium wilt using interspecific chromosome segment introgression lines
- The impacts of conservation agriculture on crop yield in China depend on specific practices,crops and cropping regions
- Exp2 polymorphisms associated with variation for fiber quality properties in cotton(Gossypium spp.)
- Isolation and characterization of a novel wall-associated kinase gene TaWAK5 in wheat(Triticum aestivum)
- Effect of subsoil tillage depth on nutrient accumulation,root distribution,and grain yield in spring maize
