Identification and Characterization of the Populus AREB/ABF Subfamily
2013-11-22LexiangJiJiaWangMeixiaYeYingLiBinGuoZhongChenHaoLiandXinminAn
Lexiang Ji,Jia Wang,Meixia Ye,Ying Li,Bin Guo,Zhong Chen,Hao Li and Xinmin An
National Engineering Laboratory for Tree Breeding,NDRC;Key Laboratory of Genetics and Breeding in Forest Trees and Ornamental Plants,MOE;The Tree and Ornamental Plant Breeding and Biotechnology Laboratory,SFA,College of Biological Sciences and Biotechnology,Beijing Forestry University,Beijing 100083,China
Introduction
Abscisic acid(ABA)is a major plant hormone that plays a fundamental role in vegetative tissue responses and adaptation to abiotic and biotic stresses,such as drought,high salinity,seed maturation,and dormancy(Bray et al.2000;Shinozaki et al.2003).However,the mechanism of ABA in cold-stressresponse gene expression remains unclear(Yamaguchi-Shinozaki and Shinozaki 2006).Previous research has suggested that genes that are induced by dehydration and cold stresses are unresponsive to exogenous application of ABA(Zhu 2002;Shinozaki et al.2003;Yamaguchi-Shinozaki and Shinozaki 2005),implying the existence of both ABA-independent and-dependent pathways.
Most ABA-induced genes contain the cis-acting ABA-responsive element(ABRE;PyACGTGGC)in their promoter regions(Bray 1994;Busk and Pages 1998).Researchers have found that ABA-responsive gene expression requires multiple ABREs or the combination of an ABRE with a coupling element(CE)as a functional promoter(Shen et al.1996;Hobo et al.1999;Narusaka et al.2003).Furthermore,ABREs control gene expression via bZIP-type ABA-responsive element binding protein/ABRE-binding factor(AREB/ABF)transcription factors(Yoshida et al.2010).The ABRE-binding(AREB)proteins,or ABRE-binding factors(ABFs),of Arabidopsis thaliana have been isolated using the yeast one-hybrid screening method(Choi et al.2000;Uno et al.2000).The ABF1 protein is induced by cold but not by osmotic stress(Kim et al.2004;Fujita et al.2005).AREB1/ABF2,AREB2/ABF4,and ABF3 are induced by dehydration,high salinity,or ABA treatment in vegetative tissue(Fujita et al.2005).These three proteins are primary transcription factors that cooperatively regulate ABRE-dependent gene expression for ABA signaling under conditions of water stress(Yoshida et al.2010).AREB3 was not detected in either stressed or unstressed plants(Uno et al.2000).
Populus trichocarpa is a species of deciduous broadleaf tree that is highly valued for conservation and timber production.P.trichocarpa also has great potential as a sustainable and renewable cellulose-based biofuel and is regarded as an alternative to fossil fuels in the future(Nieminen et al.2012).However,the growth and productivity of Populus are substantially affected by abiotic stresses,including drought,high salinity,and low temperature(Zhuang et al.2008).
The full genome sequence of P.trichocarpa was published in 2006(Tuskan et al.2006),allowing genome-wide analyses of the AREB/ABF subfamily.In the present study,we performed a comprehensive bioinformatics analysis of the Populus AREB/ABF subfamily.Tissue-specific expression profiles were investigated in eight tissue types.Furthermore,we analyzed member transcript patterns under ABA treatment.Both in silico analysis and tissue-specific and induced-expression patterns provided an integrated perspective on this protein subfamily.
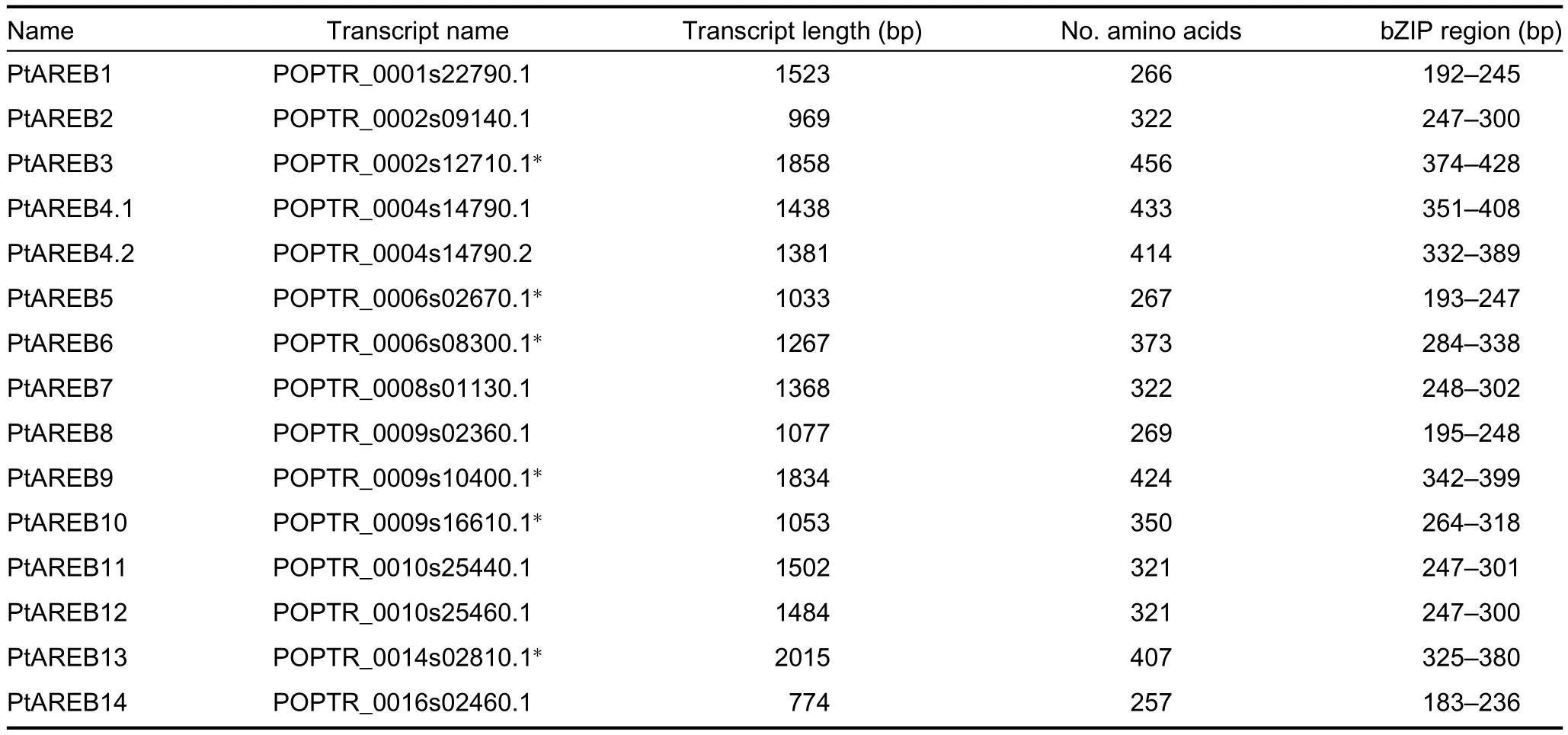
Table 1.AREB/ABF subfamily in Populus trichocarpa
Results
Isolation of the AREB/ABF subfamily in Populus trichocarpa
From the genome of A.thaliana,five AREB/ABF members were selected as bait:ABF1,AREB1/ABF2,ABF3,AREB2/ABF4,and AREB3.We applied a comprehensive two-step search process,because splicing isoforms existed at four loci.First,we used the five primary gene models(sensu TAIR)corresponding to the Arabidopsis AREB/ABF members to search within the database,and found 14 distinct genes.Second,all gene models from Arabidopsis were used in the database search query,with results similar to step one.Finally,the 14 genes were identified as putative members of the Populus AREB/ABF subfamily(Table 1).Locus POPTR_0004s14790 contained a splicing isoform that reached the E-value threshold.For more comprehensive results,we used this isoform in the subsequent analysis.Based on the functional annotations in Phytozome v8.0(Goodstein et al.2012),each of the 15 protein sequences had a basic leucine zipper(bZIP),which is characteristic of bZIP transcription factors.Six members had orthologues defined as ABA-responsive element binding factors(Table 1).
Protein structure in the Populus AREB/ABF subfamily

Multiple protein sequence alignments of the 15 amino-acid sequences were performed in BioEdit,as shown in Figure 1.The length of the aligned sequences was 526 amino acids,with 141 sites identified as relatively conserved(threshold of 50%).Thirty-eight sites were completely conserved.All five Arabidopsis AREB/ABF members contained the bZIP domain,which consisted of two motifs:a leucine zipper responsible for TF(transcription factor)dimerization and a basic region involved in the specific binding of the TF to its target DNA(Fujita et al.2011).Whole-region analysis of the 15 Populus sequences showed that each contained a bZIP domain(Figure 1,circled by a dotted line),with lengths from 54 to 58 amino acids.Within the bZIP region,39 amino acids,occupying 70.5%of the total region,were relatively conserved and 19(34.3%)were completely conserved.Our results indicate the existence of high levels of protein similarity in the Populus AREB/ABF subfamily,especially in the bZIP domain region.As previously described,the gene PtAREB4 contains a splicing isoform at the same locus(Figure 1).In comparing the two sequences,we found that 19 amino acids present in PtAREB4.1 were not translated in PtAREB4.2,suggesting that a sequence deletion occurred in the transformation of mRNA from pre-mRNA.
Phylogenetic relationships and gene structure
To verify and elucidate the relationships between P.trichocarpa and A.thaliana AREB/ABF members,we constructed a phylogenetic tree of AREB/ABF proteins.Figure 2 shows that AREB1/ABF2,ABF3,AREB2/ABF4,and ABF1 are relatively closely related compared to AREB3.Eight of the 14 Populus members were shown to have relatively high similarities to AREB3.The other six members have high genetic similarities to AREB1/ABF2.
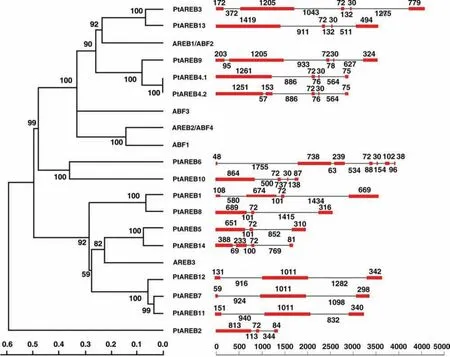
Figure 2.Neighbor-joining tree of AREB/ABF proteins from Populus trichocarpa and Arabidopsis thaliana.The gene structure of each AREB/ABF gene in P.trichocarpa is shown.The blocks and lines represent exons and introns,respectively.The numbers above and below the exons and intron are sizes(bp).
To further classify and analyze the relationships of the Populus AREB/ABF subfamily,we constructed gene structures for this subfamily(Figure 2).The genomic sequence lengths ranged from 1,426 to 4,680 base pairs(bp),with a maximum of seven exons in PtAREB6 and a minimum of three exons in PtAREB3.This indicated high variability of sequence lengths within this subfamily.A combined analysis of the phylogenetic linkage and gene structure indicated a homologous relationship.Three member proteins,PtAREB7,PtAREB11,and PtAREB12,were closely related on the tree.Furthermore,these three genes had similar structures to four of their exons in the gene sequences.Their genes were short,ranging from 3,274 to 3,682 bp,with each of their third exons being 1,011 bp long.The 11 other genes all contained an exon that was 72 bp long.Six of the genes had a 30-bp exon adjacent to the 72-bp exon.
Locations of the Populus AREB/ABF subfamily
All 14 Populus AREB/ABF genes were located on nine of the 19 chromosome scaffolds(Figure 3).Four scaffolds contained more than one locus,with PtAREB2 and PtAREB3 on Scaffold 2,PtAREB5 and PtAREB6 on Scaffold 6,PtAREB8,PtAREB9,and PtAREB10 on Scaffold 9,and PtAREB11 and PtAREB12 on Scaffold 10.The two genes on Scaffold 10 were relatively closely related and had a similar gene structure(Figure 2).
Expression patterns of the AREB/ABF genes
To identify the tissue-specific expression profiles of the Populus AREB/ABF subfamily,we performed qRT-PCR on eight different tissue types for 14 selected genes(Figure 4).Because the transcript levels of the 14 Populus AREB members varied so widely relative to each other,they were graphed on different scales.Based on normalized gene expression levels,young and mature leaf tissues had the most abundant transcripts of eleven genes(PtAREB1,3,4,5,6,7,9,10,11,12,and 13),while root tissue had the highest transcript levels of two genes(PtAREB8 and 14).Previous studies indicated that ABA regulates many crucial processes,including control of stomatal closure and adaptive responses to various abiotic stresses(Finkelstein et al.2002;Yamaguchi-Shinozaki and Shinozaki 2006).These tissue-specific patterns of Populus AREB members agree with the known functions of ABA.Three genes(PtAREB6,10,and 14)were detected in some,but not all,of the eight tissue types,and one additional gene(PtAREB8)had relatively weak transcript levels.In contrast,PtAREB13 had much higher expression levels than all other subfamily members in young and mature leaves.
With exogenous application of ABA,eight genes(PtAREB2,3,5,6,7,8,10,and 14)were upregulated;the expressions of seven of them reached maximum levels after 12 h,then gradually decreased(Figure 5).Three members,PtAREB1,PtAREB11,and PtAREB12,were constantly downregulated under ABA stress,while the expressions of PtAREB4,PtAREB9,and PtAREB13 decreased rapidly during the initial 4 h,then gradually increased.
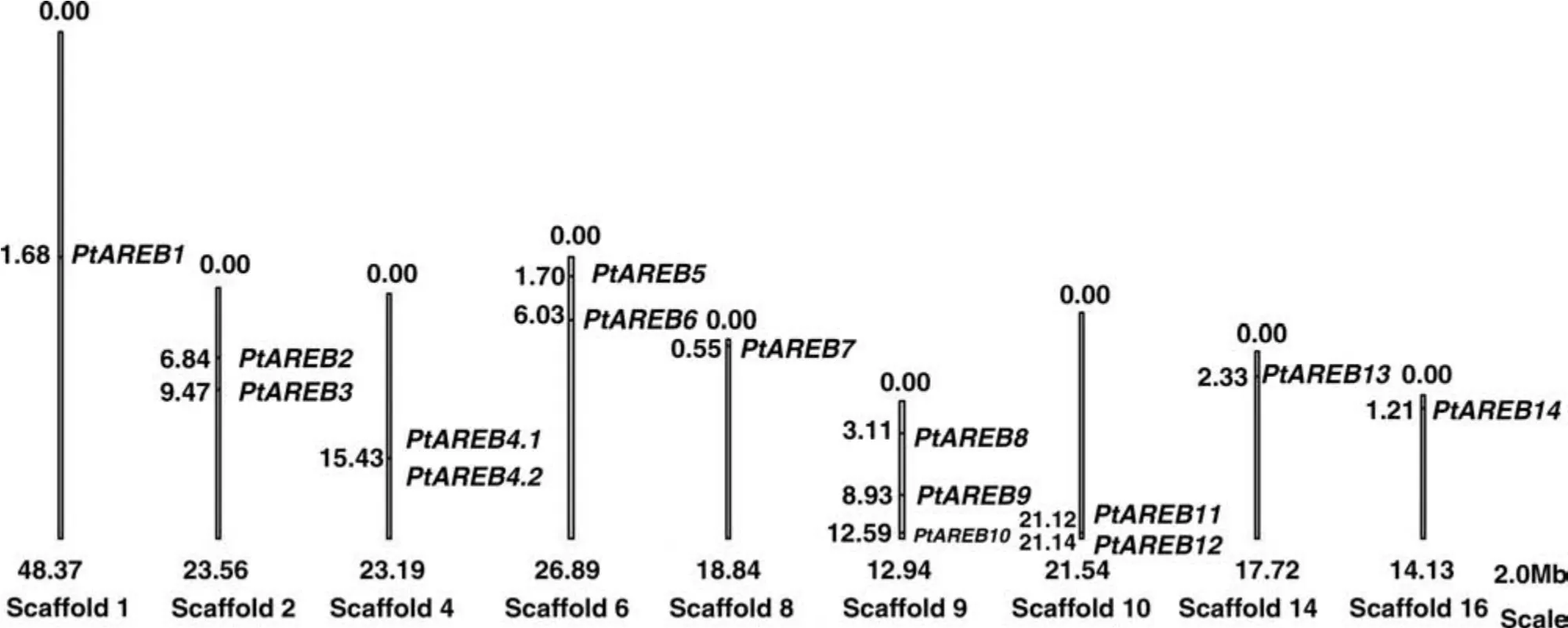
Figure 3.Physical locations of Populus AREB/ABF genes on the scaffolds.The scale bar represents 2.0 Mb.Scaffold numbers and sizes(Mb)are indicated below each scaffold.
Discussion
The first complete plant genome sequenced was that of A.thaliana,which has one of the smallest plant genomes,with about 125 Mb pairs and five chromosomes(Swarbreck et al.2008).In contrast,the P.trichocarpa genome has about 485 Mb pairs and 19 chromosomes(Tuskan et al.2006),and is four times larger than that of Arabidopsis.To avoid false positives in the identification of putative members of the Populus AREB/ABF subfamily,we used an E-value threshold of 1e-30.This E value was small compared with previous studies on P.trichocarpa,such as those by Zhuang et al.(2008)and Shao et al.(2011).Only 14 genes met this threshold;all of them contained bZIP domains and were either upregulated or downregulated under ABA stress.We propose that the combination of in silico screens and qRT-PCR is a feasible method for genome-wide analysis of the Populus AREB/ABF subfamily.
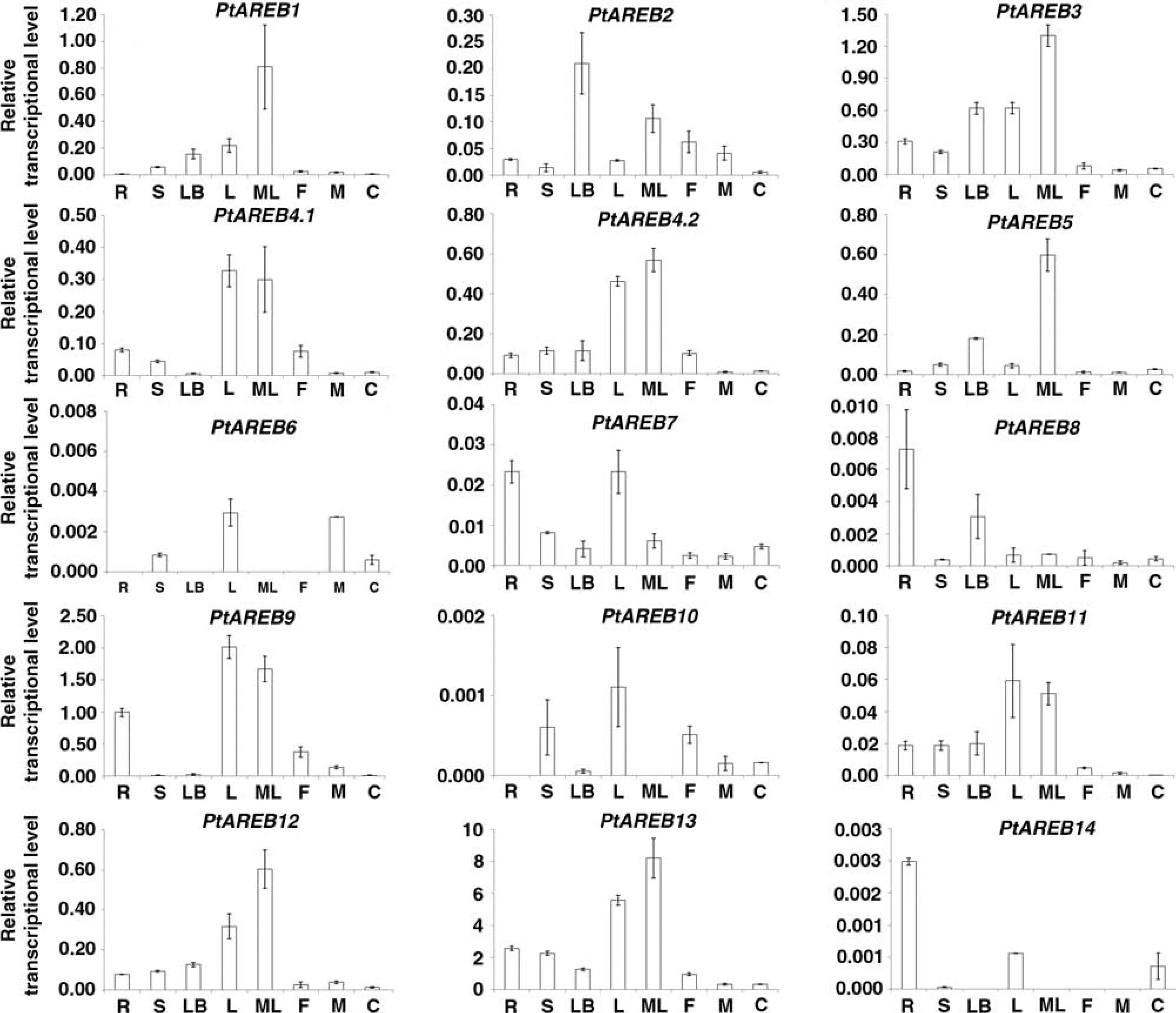
Figure 4.Expression analyses of 14 Populus AREB/ABF genes using qRT-PCR.The endogenous reference gene was poplar ACTIN.Error bars represent the standard deviation(SD)of three technical repeats.Letters on the x-axis are:R,root;S,stem;LB,leaf bud;L,leaf;ML,mature leaf;F,female floral bud;M,male floral bud;C,female catkin.
Gene duplication is common in biological evolution.Previous reports showed that many mutations in functional genes had little or no phenotypic effects,suggesting that these mutations could be partly or completely compensated for by the activity of other genes;this process is called redundant genetic function(Lambie and Kimble 1991;Pickett and Meeks-Wagner 1995).Genes within a family of duplicates may have similar structures and functions,and partial redundancy is often seen among gene family members(Pickett and Meeks-Wagner 1995;Shao et al.2011).In this study,four scaffolds each contained more than one member of the AREB/ABF subfamily.Scaffolds 2,6,and 9 each contained AREB/ABF genes with partially or completely different gene structures and amino acid sequences,as well as different tissue-specific expression patterns.However,all of those members were upregulated under ABA stress except PtAREB9.On Scaffold 10,PtAREB11 and PtAREB12 had a relatively close phylogenetic relationship and similar gene structures;they also showed similar expression profiles in all eight tissues,and their expression levels decreased with the application of exogenous ABA.Only three genes were constantly suppressed under ABA stress.These data suggest that PtAREB11 and PtAREB12 may share some overlapping functions and compensate for one another.
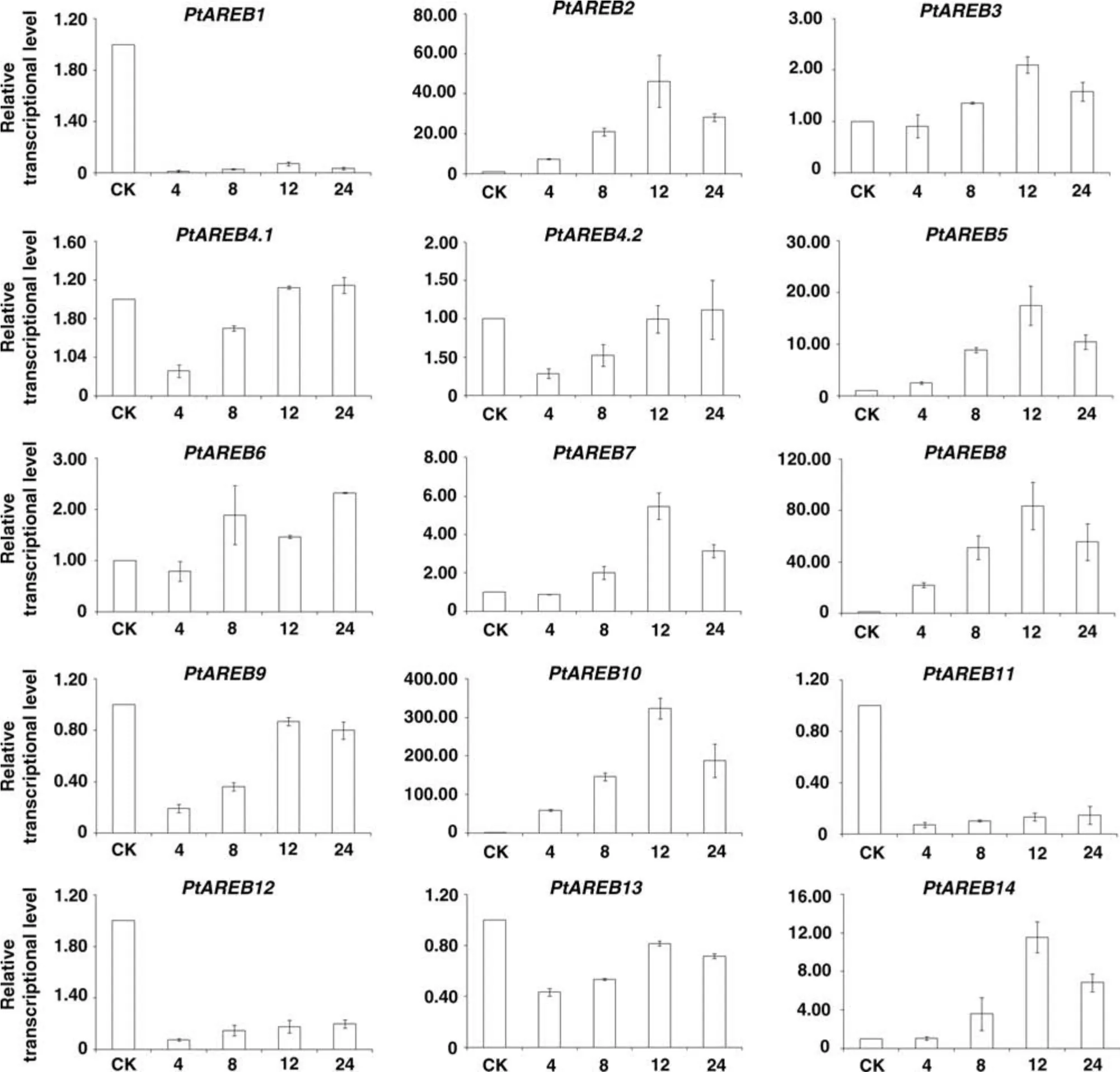
Figure 5.Expression profiles of Populus AREB/ABF members under Abscisic Acid(ABA)stress.The leaves were collected after 0(CK),4,8,12,or 24 h of exogenous 100µM ABA treatment.Poplar ACTIN was used as an endogenous reference gene.Error bars represent the standard deviation(SD)of three technical repeats.The normalized mRNA levels at 0 h were arbitrarily set to 1.
The AREB3 gene was first cloned using yeast one-hybrid screening with the RD29B promoter,and it was undetected in both unstressed and stressed plants(Uno et al.2000).Of the 75 members of the bZIP family in the Arabidopsis genome,nine are considered to be involved in ABA signaling(Bensmihen et al.2002).Based on phylogenetic relationships and functional analyses,AREB3 was classified into the ABI5/AtDPBF subfamily,which also includes ABI5,EEL,DPBF2/AtbZIP67,and DPBF4.This family is mainly expressed in seeds and appears to play vital roles in seed maturation and development(Fujita et al.2011).In this study,we found that eight Populus AREB/ABF members were relatively closely related to AREB3.Interestingly,none of them were expressed like AREB3 in A.thaliana.All were detected in unstressed tissues;four were upregulated and four were downregulated by the ABA treatment.Hoth et al.(2010)identified the relatively closely related AREB3 and ABI5 as potential interactors.Other researchers also suggested that ABI5 was upregulated under abiotic stresses(Nakashima et al.2006).These cited studies may partially explain the expression profiles of the Populus AREB/ABF members.
The other four AREB/ABF members,including AREB1/ABF2,AREB2/ABF4,ABF1,and ABF3,are mainly expressed in vegetative tissues under abiotic stress conditions(Fujita et al.2011).ABF1 is not induced by osmotic stress,including ABA treatment(Fujita et al.2005).In this study,all 14 AREB/ABF members were either enhanced or repressed by exogenous ABA.The responses of these genes to ABA may be species-specific and may imply complex ABA transcriptional regulatory networks in Populus.
Many abiotic stress-related transcription factors can be activated or repressed by WRKY proteins.The AtWRKY40-ABI5-AtWRKY63 module is part of the ABA-signaling network.In this module,AtWRKY40 is upstream of ABI5,and it represses the expression of ABI5 under unstressed conditions.As ABA accumulates,AtWRKY40 moves out of the nucleus,which de-represses ABI5 at the transcriptional level.Activation of ABI5 induces the downstream AtWRKY63,which then activates further downstream target genes such RD29A and ABF2(Rushton et al.2012).In this study,we found that six AREB/ABF members were downregulated under ABA stress,and three were constantly repressed under the stress.They may act as the upstream genes that repress ABA-induced genes in unstressed conditions,and may de-repress with when ABA is perceived by receptors.
In summary,we performed a relatively complete bioinformatics analysis of the Populus AREB/ABF subfamily.We identified 14 genes as putative AREB/ABF genes and assessed their expressions in eight different tissue types via qRT-PCR.Eight were upregulated by ABA treatment,whereas six decreased in expression under the stress.These data indicated the complexity of Populus ABA-mediated transcriptional regulation networks.Our results will help elucidate the functions of the Populus AREB/ABF subfamily,providing clues for the identification of candidate genes involved in abiotic stress responses in plants.
Materials and Methods
Database search and sequence retrieval
We obtained the Arabidopsis thaliana L.AREB/ABF sequences from The Arabidopsis Information Resource(TAIR)(Swarbreck et al.2008).To anchor the Populus AREB/ABF subfamily,the amino acid sequences of the Arabidopsis AREB/ABF members were used in our BLAST search using the Joint Genome Institute(JGI)Phytozome portal(Goodstein et al.2012).This search enabled us to identify sequence similarities using the latest(v2.2)JGI gene annotation of the v.2 Populus trichocarpa Torr.&Gray.assembly.We applied an E-value of 1e-30 to reduce false positives.
Construction of the protein alignment
The amino acid sequences retrieved from Phytozome were aligned using BioEdit 7.0.9 software(Hall 1999).We used a 50%threshold to define the relatively conserved regions of the amino-acid sequence.
Phylogenetic tree and gene structure
We constructed a neighbor-joining(NJ)tree with genetic distance matrices of protein sequences using the Poisson model,pairwise deletion,and the bootstrap method with 1000 replications in MEGA v.5.0 software(Tamura et al.2011).We used exon/intron data to build the gene structure for this study.
Physical position of the AREB/ABF genes
The physical locations of Populus AREB/ABF subfamily were analyzed using the JGI Populus v2.2 gene annotation.This annotation covered 403 Mb-pairs of the genome with an average assembled read depth of 7.45×,and assembled into 2,518 scaffolds(Goodstein et al.2012).The first 19 scaffolds from the assembly corresponded to the 19 poplar chromosomes that were formerly used to describe the P.trichocarpa genome(Tuskan et al.2006).However,more than 2,000 scaffolds remained unassembled to corresponding chromosomes;therefore,to characterize the physical locations of the genes,we used scaffolds rather than chromosomes,as described in Phytozome.
Plant materials and treatment
We selected eight different tissue types from Populus tomentosa Carr.for analysis.We collected female catkins,female floral buds,male floral buds,vegetative buds,and mature leaves from adult female(5082)and male(LM50)trees growing in the Beijing Forestry University nursery.Tissues were immediately placed in liquid nitrogen and stored at-80°C until RNA extraction.We collected samples of root,stem,and young leaf tissue from one-m-old tissue-cultured plantlets and extracted total RNA immediately after sampling.To test the effects of ABA treatment,roots of one-m-old tissue-cultured plantlets were submerged in a 100µM ABA solution(Choi et al.2000;Fujita et al.2005;Shao et al.2011),and leaves were harvested for immediate RNA extraction after 0,4,8,12,and 24 h.
RNA isolation and qRT-PCR
Total RNAs were extracted from combined samples of each tissue from multiple individuals using a modified CTAB method(Chang et al.1993).The mRNA was then pretreated with RQ1 DNase(Promega,Madison,WI,USA)to remove genomic DNA contaminants.Subsequently,first-strand cDNA was synthesized using mRNA as a template using the Reverse Transcription System(Promega).
Quantitative real-time PCR(qRT-PCR)was performed in an ABI PRISM 7500 Fast Real-time PCR System(Applied Biosystems,Foster City,CA,USA)using the SYBR Premix Ex TaqTMKit(TaKaRa,Kyoto,Japan)with amplification conditions as recommended by TaKaRa.All reactions were run in triplicate for each sample,and the poplar ACTIN gene(Accession:AY261523.1)was selected as the endogenous control gene for normalization(An et al.2011).All primers are listed in Table S1.
This work was supported by the Major State Basic Research Development Program(2012CB114505),the National Natural Science Foundation of China(31170631),and the National Hightech Research and Development Program(2011AA100201)of China.
An X,Ye M,Wang D,Wang Z,Cao G,Zheng H,Zhang Z(2011)Ectopic expression of a poplar APETALA3-like gene in tobacco causes early flowering and fast growth.Biotechnol.Lett.33,1239–1247.
Bensmihen S,Rippa S,Lambert G,Jublot D,Pautot V,Granier F,Giraudat J,Parcy F(2002)The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis.Plant Cell 14,1391–1403.
Bray EA(1994)Alterations in gene expression in response to water deficit.In:Basra AS,ed.Stress-Induced Gene Expression in Plants.Amsterdam:273Harwood Academic,India.pp.1–23.
Bray EA,Bailey-Serres J,Weretilnyk E(2000)Responses to abiotic stresses.In:Gruissem W,Buchannan B,Jones R,eds.Biochemistry and Molecular Biology of Plants.Amer.Soc.Plant Physiol.,Rockville,MD.pp.1158–1203.
Busk PK,Pages M(1998)Regulation of abscisic acid-induced transcription.Plant Mol.Biol.37,425–435.
Chang S,Puryear J,Cairney J(1993)A simple and efficient method for isolating RNA from pine trees.Plant Mol.Biol.Rep.11,113–116.
Choi H,Hong J,Ha J,Kang J,Kim SY(2000)ABFs,a family of ABA-responsive element binding factors.J.Biol.Chem.275,1723–1730.
Finkelstein RR,Gampala SS,Rock CD(2002)Abscisic acid signaling in seeds and seedlings.Plant Cell 14 Suppl,S15–45.
Fujita Y,Fujita M,Satoh R,Maruyama K,Parvez MM,Seki M,Hiratsu K,Ohme-Takagi M,Shinozaki K,Yamaguchi-Shinozaki K(2005)AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis.Plant Cell 17,3470–3488.
Fujita Y,Fujita M,Shinozaki K,Yamaguchi-Shinozaki K(2011)ABA-mediated transcriptional regulation in response to osmotic stress in plants.J.Plant Res.124,509–525.
Goodstein DM,Shu S,Howson R,Neupane R,Hayes RD,Fazo J,Mitros T,Dirks W,Hellsten U,Putnam N,Rokhsar DS(2012)Phytozome:A comparative platform for green plant genomics.Nucleic Acids Res.40,D1178–1186.
Hall TA(1999)BioEdit:A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT.Nucl.Acids Symp.Ser.41,95–98.
Hobo T,Kowyama Y,Hattori T(1999)A bZIP factor,TRAB1,interacts with VP1 and mediates abscisic acid-induced transcription.Proc.Natl.Acad.Sci.USA 96,15348–15353.
Hoth S,Niedermeier M,Feuerstein A,Hornig J,Sauer N(2010)An ABA-responsive element in the AtSUC1 promoter is involved in the regulation of AtSUC1 expression.Planta 232,911–923.
Kim S,Kang JY,Cho DI,Park JH,Kim SY(2004)ABF2,an ABRE-binding bZIP factor,is an essential component of glucose signaling and its overexpression affects multiple stress tolerance.Plant J.40,75–87.
Lambie EJ,Kimble J(1991)Two homologous regulatory genes,lin-12 and glp-1,have overlapping functions.Development 112,231–240.
Nakashima K,Fujita Y,Katsura K,Maruyama K,Narusaka Y,Seki M,Shinozaki K,Yamaguchi-Shinozaki K(2006)Transcriptional regulation of ABI3-and ABA-responsive genes including RD29B and RD29A in seeds,germinating embryos,and seedlings of Arabidopsis.Plant Mol.Biol.60,51–68.
Narusaka Y,Nakashima K,Shinwari ZK,Sakuma Y,Furihata T,Abe H,Narusaka M,Shinozaki K,Yamaguchi-Shinozaki K(2003)Interaction between two cis-acting elements,ABRE and DRE,in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses.Plant J.34,137–148.
Nieminen K,Robischon M,Immanen J,Helariutta Y(2012)Towards optimizing wood development in bioenergy trees.New Phytol.194,46–53.
Pickett FB,Meeks-Wagner DR(1995)Seeing double:Appreciating genetic redundancy.Plant Cell 7,1347–1356.
Rushton DL,Tripathi P,Rabara RC,Lin J,Ringler P,Boken AK,Langum TJ,Smidt L,Boomsma DD,Emme NJ,Chen X,Finer JJ,Shen QJ,Rushton PJ(2012)WRKY transcription factors:Key components in abscisic acid signalling.Plant Biotechnol.J.10,2–11.
Shao Y,Wei G,Wang L,Dong Q,Zhao Y,Chen B,Xiang Y(2011)Genome-wide analysis of BURP domain-containing genes in Populus trichocarpa.J.Integr.Plant Biol.53,743–755.
Shen Q,Zhang P,Ho TH(1996)Modular nature of abscisic acid(ABA)response complexes:Composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley.Plant Cell 8,1107–1119.
Shinozaki K,Yamaguchi-Shinozaki K,Seki M(2003)Regulatory network of gene expression in the drought and cold stress responses.Curr.Opin.Plant Biol.6,410–417.
Swarbreck D,Wilks C,Lamesch P,Berardini TZ,Garcia-Hernandez M,Foerster H,Li D,Meyer T,Muller R,Ploetz L,Radenbaugh A,Singh S,Swing V,Tissier C,Zhang P,Huala E(2008)The Arabidopsis Information Resource(TAIR):Gene structure and function annotation.Nucleic Acids Res.36,D1009–1014.
Tamura K,Peterson D,Peterson N,Stecher G,Nei M,Kumar S(2011)MEGA5:Molecular evolutionary genetics analysis using maximum likelihood,evolutionary distance,and maximum parsimony methods.Mol.Biol.Evol.28,2731–2739.
Tuskan GA,Difazio S,Jansson S,Bohlmann J,Grigoriev I,Hellsten U,Putnam N,Ralph S,Rombauts S,Salamov A,Schein J,Sterck L,Aerts A,Bhalerao RR,Bhalerao RP,Blaudez D,Boerjan W,Brun A,Brunner A,Busov V,Campbell M,Carlson J,Chalot M,Chapman J,Chen GL,Cooper D,Coutinho PM,Couturier J,Covert S,Cronk Q,Cunningham R,Davis J,Degroeve S,Dejardin A,Depamphilis C,Detter J,Dirks B,Dubchak I,Duplessis S,Ehlting J,Ellis B,Gendler K,Goodstein D,Gribskov M,Grimwood J,Groover A,Gunter L,Hamberger B,Heinze B,Helariutta Y,Henrissat B,Holligan D,Holt R,Huang W,Islam-Faridi N,Jones S,Jones-Rhoades M,Jorgensen R,Joshi C,Kangasjarvi J,Karlsson J,Kelleher C,Kirkpatrick R,Kirst M,Kohler A,Kalluri U,Larimer F,Leebens-Mack J,Leple JC,Locascio P,Lou Y,Lucas S,Martin F,Montanini B,Napoli C,Nelson DR,Nelson C,Nieminen K,Nilsson O,Pereda V,Peter G,Philippe R,Pilate G,Poliakov A,Razumovskaya J,Richardson P,Rinaldi C,Ritland K,Rouze P,Ryaboy D,Schmutz J,Schrader J,Segerman B,Shin H,Siddiqui A,Sterky F,Terry A,Tsai CJ,Uberbacher E,Unneberg P,Vahala J,Wall K,Wessler S,Yang G,Yin T,Douglas C,Marra M,Sandberg G,Van de Peer Y,Rokhsar D(2006)The genome of black cottonwood,Populus trichocarpa(Torr.&Gray).Science 313,1596–1604.
Uno Y,Furihata T,Abe H,Yoshida R,Shinozaki K,Yamaguchi-Shinozaki K(2000)Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions.Proc.Natl.Acad.Sci.USA 97,11632–11637.
Yamaguchi-Shinozaki K,Shinozaki K(2005)Organization of cisacting regulatory elements in osmotic-and cold-stress-responsive promoters.Trends Plant Sci.10,88–94.
Yamaguchi-Shinozaki K,Shinozaki K(2006)Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses.Annu.Rev.Plant Biol.57,781–803.
Yoshida T,Fujita Y,Sayama H,Kidokoro S,Maruyama K,Mizoi J,Shinozaki K,Yamaguchi-Shinozaki K(2010)AREB1,AREB2,and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation.Plant J.61,672–685.
Zhu JK(2002)Salt and drought stress signal transduction in plants.Annu.Rev.Plant Biol.53,247–273.
Zhuang J,Cai B,Peng RH,Zhu B,Jin XF,Xue Y,Gao F,Fu XY,Tian YS,Zhao W,Qiao YS,Zhang Z,Xiong AS,Yao QH(2008)Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa.Biochem.Biophys.Res.Commun.371,468–474.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1.Primers used for qRT-PCR analysis.
