Lung Cancer: MicroRNA and Target Database
2012-09-11ChallaKIRNPonnalaDEEPIKA
Challa KIRN, Ponnala DEEPIKA
Centre for Cellular and Molecular Biology, India
Abstract MicroRNAs (miRNAs) are a class of non-coding RNAs that hybridize to mRNAs and induce either translation repression or mRNA cleavage. Recently, it has been reported that miRNAs could possibly play a critical role in cellular processes like regulation of cell growth, differentiation, and apoptosis, emphasizing their role in tumorigenesis. Likewise, several miRNA’s are involved in lung cancer tumorigenesis. The present review puts forth a database of human miRNA’s involved in lung cancer along with their target genes. It also provides sequences of miRNA’s and their chromosomal locations retrieved from different databases like microCosm (218 microRNAs), PhenomiR (293 microRNAs), and mir2Disease (90 microRNAs) and target gene information such as the pathways like cell cycle regulation, angiogenesis, apoptosis etc. Though miRNA’s are still to be explored, they hold a promise as therapeutic targets and diagnostic markers of cancer.
Key words Lung neoplasms; MicroRNAs; Targets
Introduction
MicroRNAs (miRNA) are short, noncoding RNAs regulating gene expression[1,2]. MicroRNAs were discovered in 1993 by Victor Ambros and group. However, their importance as an abundant class of regulatory non-coding RNAs came into light only after their rediscovery by Reinhart[3]. MicroRNAs are reported to cover 1% to 3% of the genomes and 30% of the genes are shown to be regulated by them (Lai[4]). Thousands of miRNAs involved in various human diseases have been identified by Jin[5]. The reason for their involvement is mainly due to a change in the copy numbers or due to mutations in the sequence of miRNA or their target sites[6]. They were also shown to be deregulated in majority of cancers[7]. Recent studies have demonstrated the critical role of miRNAs in cancer pathogenesis[8-14].The potential of restoring levels of aberrantly under expressed miRNAs with miRNA mimics or inactivating over-expressed miRNAs with miRNA inhibitors has been explored and shows promise as the next generation of therapeutic strategies[15-19].
Lung cancer is one of the most common causes of cancer related deaths in the world and is responsible for 1.3 million deaths worldwide every year. Lung cancer can be divided into two groups according to pathology: non-small cell lung cancer (NSCLC) (80.4%) and small cell lung cancer(SCLC) (16.8%)[20]. Many factors potentially contribute to lung cancer formation for e.g. tobacco smoke, ionizing radiations and viral infections. However, the mechanisms involved in lung carcinogenesis remain largely unknown.Lung cancer is initiated by activation of oncogenes or inactivation of tumor suppressor genes[21]. Similar to other cancers, many pathways are involved in lung cancer development and progression.
MicroRNAs are involved in many pathways related to lung cancer. ErbB2 (HER2/Neu), ErbB3 and ErbB4 belong to the family of receptor tyrosine kinases. These kinases initiate signal cascades leading to DNA synthesis and cell proliferation, and their over-expression is often critically involved in tumorigenesis and cancer cell proliferation[22]. The epidermal growth factor receptor (EGFR)/ErbB1 regulates cell proliferation, apoptosis, angiogenesis and tumor invasion in non-small cell lung carcinoma[21]. Recent investigations have demonstrated that miRNAs are likely to be involved in the regulation of EGFR in lung cancer. An inhibitor of miR-128b resulted in up-regulated EGFR expression in an EGFR-expressing NSCLC cell line, and treatment with a miR-128b mimic resulted in a concomitant reduction of EGFR expression[23]. Inhibition of miR-21 was recently shown to enhance the anti-apoptotic potential of an anti-EGFR tyrosine kinase inhibitor in an EGFR-mutant lung adenocarcinoma cell line[24]. The proto-oncogene RAS is the central molecule of the SOS-RS-Raf-MAPK cascade which is the downstream cytoplasmic effector of the growth factor receptors. Approximately 20% of tumors in general and 30% of lung tumors have activating mutations in one of the RAS genes[25]. One of the downstream effectors of the RAS signaling pathway is the MYC oncogene. MYC amplif i cation and over-expression have been detected in different histologic subtypes of lung cancer[26]. Both RS and MYC are targets of the let-7/miR-98 family of miRNAs. Johnson[27]showed that members of the let-7 family of miRNAs bind to the 3′UTR of RS, down-regulating RS expression in human cells. It is seen that let-7 is poorly expressed in lung cancer compared to normal lung tissue, and that the expression of let-7 is inversely correlated with the expression of RAS in lung tumor samples[28]. miR-29 is down-regulated in a number of human cancers including lung cancers, and its ability to repress Mcl-1 coupled with its role in regulating epigenetic DNA methylation[29]means that enhancing expression of this regulatory element could be an effective therapeutic strategy[30-32]. Vascular endothelial growth factor (VEGF)is an important mediator of angiogenesis during tumor development, and this molecule and its receptor (VEGFR)have been primary targets of therapies designed to target pathological angiogenic signaling[33]. Liu[34]investigated the involvement of miR-126 in the regulation of angiogenic processes in a lung cancer model. They found decreased expression of miR-126 and increased expression of VEGF-A in various lung cancer cell lines and showed that introduction of miR-126 using a lentiviral vector could down-regulate the expression of VEGF-A and inhibit growth.
Inactivation of tumor suppressor genes plays important role in lung carcinogenesis. The p53 tumor suppressor gene, located on chromosome 17p, is affected in 60%-75%of lung cancer including both NSCLC and SCLC while Rb is more likely inactivated in SCLC[35]. In lung cancer cells,induction of miR-34 results in apoptosis[36,37]and miRNA profiling shows that the expression of miR-34a, miR-34b,and miR-34c are directly correlated with expression of the p53 tumor suppressor[38], suggesting that miR-34 is involved in regulating apoptosis as a regulatory target of p53. Evidence is emerging that tumor suppressors are likely to be regulated by miRNA activity. FUS1/TUSC2 is a tumor suppressor gene located on 3p21.3 that has been shown to be negatively regulated by the activities of miR-197,miR- 93, miR-98, and miR-378[13,39]. Reduced or complete loss of Fus1 expression was found in 82% and 100% of non-small cell and small cell lung cancer cell lines[40], and elevated levels of miR-93 and miR-197 have been shown to correlate with reduced Fus1 expression in NSCLC tumor specimens[39]. Another important tumor suppressor gene is LKB1, whose loss-of-function mutation/deletion is observed in 30% lung adenocarcinomas and 20% of squamous cell carcinomas[41,42]. An investigation of miR-126 in the regulation of invasive potential in lung cancer revealed an inverse relationship between Crk and miR-126 expression in squamous cell tumors[43]. miR-21 also targets PDCD4, a proapoptotic gene that inhibits tumorigenesis and whose downregulation has been linked to poor survival in colon and lung cancer patients[44-46]. The let-7 family has been shown to inhibit the expression of several oncogenes including RS,MYC, and HMGA2[28,47-49]. Several studies have implicated the miR-17-92 cluster of miRNAs as an actual oncogene with important regulatory effects on pathologic tumor cell proliferation in various tissues including breast and lung[50-52].
Lung cancer microRNAs database
All these genetic pathways play a critical role in lung cancer progression. MicroRNAs play a very important role in the regulation of these pathways. Finding the targets of miRNA is equally important as finding the miRNA itself[53]. Identifying these miRNAs and their target genes provides a platform to work towards the understanding of lung cancer pathogenesis. Computational prediction of target genes for miRNAs in animals is a challenging task for both experimental and computational group, due to the complexity of miRNA target recognition[54].
Numerous databases are available which facilitate easy and efficient mining of microRNAs and their target genes involved in a specif i c cancer. Databases based on sequence annotation and microRNA genomics are miRBase[55],human microRNA disease database[56], miRNAMap[57],and miRGen[58]. miRBase serves as a central database for experimentally supported mature miRNA sequences for each supported miRNA. It also provides the genomic coordinates of the predicted precursor sequences. TargetScan[59],PicTar[60], TargetMiner[61], miRDB are the databases for miRNA target prediction and functional annotations in animals[62]. microRNA.org target and expression 2005, are the databases used for microRNA target predictions based on the complementary perfect base-pairing in the seed region of the targets. TarBase[63]is a resource of experimentally validated microRNA targets.
However, no database exists which provides a complete overview of microRNAs controlling lung cancer pathogenesis and progression. This review puts forth a database including almost all the microRNAs involved in lung carcinogenesis.The miRNAs involved in lung cancer are pooled up from databases, MicroCosm (218 microRNAs), PhenomiR(293 microRNAs) and mir2Disease (90 microRNAs). A supplementary excel document is provided with this review which gives a detailed description of the MiRNA’s involved in lung cancer. It also includes information about the accession numbers, chromosomal positions and validated targets of the respective microRNA’s.
MicroRNAs involved in lung cancer were derived from miRBase, PhenomiR[64], MiR2Disease database[56]. After manually curating the miRNAs from different databases accession numbers and sequences of microRNAs were collected from the miRBase database. MicroRNAs chromosome positions were obtained from the HUGO gene nomenclature committee (HGNC). PhenomiR database contains the microRNA expression levels in the different lung cancer samples and cell lines, those were collected for specific microRNAs which are present in PhenomiR database and related to lung cancer.
Experimentally validated target interactions
Experimentally validated targets for these lung cancer microRNAs were collected from the mir2disease and TarBase databases. These target genes were submitted to gene Go analysis software. Gene Go is the pathway analysis software which gives the information about interaction between the different biological molecules.Many target genes were found to be involved in lung cancer development and progression and these proteins are present in extracellular, membrane, cytoplasm and nucleus. All these target genes are positively or negatively regulated by the each other. Receptor ligand, generic kinase, transcription factors, lipid phosphatase, generic phosphatase, generic binding proteins and RAS super family proteins have been found to be involved in the lung cancer development. Generic binding proteins like cyclin D1, P27KIP1, and BCL2 are very important molecules in the cytoplasmic region and regulate many other targets.C-Myc is one of the important transcription factor which is present in the nucleus and regulates many proteins in the cytoplasmic region. Interactions between the extracellular,membranes, cytoplasmic and nuclear proteins are shown in the Fig 1.
The major pathways associated in lung cancer based on genego analysis are pathways involving cell proliferation,cell cycle regulation, angiogenesis, signal transduction and apoptosis. The signif i cant protein found to regulate most of the other proteins is c-Myc and the proteins regulated by c-Myc are cyclins, cyclin dependant kinases (CDKs), DNA methyl transferases, and CDK inhibitors. C-Myc is a basichelix-loop-helix/leucine zipper (bHLH/LZ) transcription factor that controls the G1-S cell-cycle transition and is over expressed in many human tumors. A thorough analysis of the pathway circuit produced by genego software depicts that C-myc directly or indirectly regulates most of the proteins which involve in various other pathways. Thus c-Myc is a potent protein which needs to be studied.
It is also seen that p27KIP1 too regulates quite a few proteins. p27KIP1 is a cell-cycle regulatory protein that interacts with cyclin-CDK2 and -CDK4, inhibiting cell cycle progression at G1. But an altered expression of p27KIP1 dysregulates the expression of proteins involved in cell cycle regulation and thereby leads to cell proliferation.Interestingly, the pathway circuit obtained from genego software reveals that c-Myc indirectly regulates p27KIP1 through Bcl2 and H-Ras. Hence c-Myc becomes further more a vital protein to be focused on, apart from p27KIP1.
MicroRNA regulation of lung cancer-related pathways provides numerous opportunities for therapeutic intervention. Competitive inhibition of miRNAs or the delivery of exogenously produced miRNAs could provide substantial therapeutic benefit by reducing the activity of prooncogenic miRNAs. These approaches are still in the preclinical stages, although intriguing evidence is emerging that supports their use in clinical applications. Expression of miR-29 family members has been shown to reduce tumorigenic potential in a lung cancer model[29]. Exogenous delivery of a synthetic let-7 mimic has been used to mediate remission of established NSCLC tumors in mice[28]. Future work will focus on both chemical modif i cations and delivery vehicles in order to maximize the therapeutic benefits and minimize the side effects of strategies designed to alter intracellular levels of specif i c miRNAs (Fig 2).
Conclusion
The study of microRNAs has evolved rapidly since the discovery of this important class of regulatory molecules.The proposed lung cancer microRNA database would prove to be helpful to lung cancer biologists in one or the other way. Further studies on the miRNA’s listed in the database would explore and explain the relation between miRNAs and the initiation and progression of malignant neoplasias and the basic biological roles that they play in tumor cell survival, invasion, metastasis, and angiogenesis. However,many issues are still to be addressed before the miRNAs can be used as therapeutic tools in the fi eld of oncology. miRNA dysregulation in particular tumor subtypes need to be further investigated, which may aid the development of new therapies that specif i cally target tumor cells with particular molecular and regulatory profiles. Overall, miRNAs are ubiquitous regulatory molecules that hold great promise for increasing our understanding of lung cancer and for improving upon current diagnostic and therapeutic methods.Further evaluation of their value in cancer diagnostics and of the functional consequences of their manipulation will provide researchers and oncologists with a wealth of new tools to characterize and treat this prevalent and diverse disease.
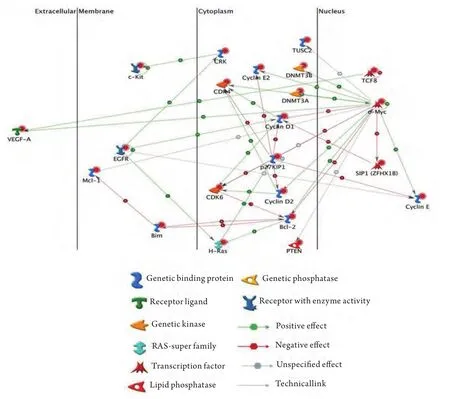
Fig 1 Lung cancer target interactions between the extracellular, membranes, cytoplasmic and nuclear proteins
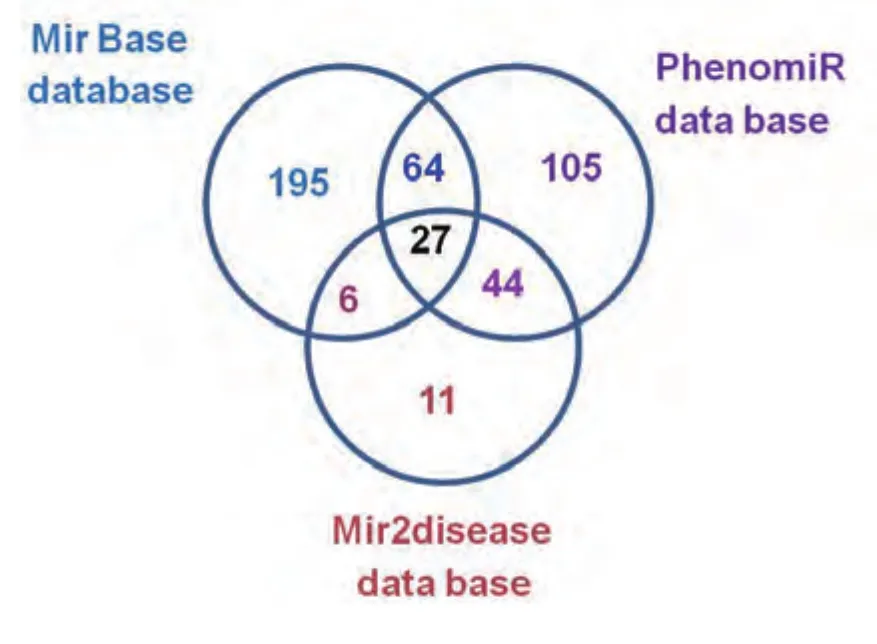
Fig 2 Venn diagram showing the distribution of total microRNAs identified form different databases. The numbers indicate the total microRNAs identified from each category and the number of microRNAs commonly identified between them.
