含2-氨基吡啶及去甲基斑蝥酸根的钴(Ⅱ)配合物的晶体结构、与DNA和BSA相互作用及体外抗增殖活性
2012-09-09张帆林秋月郑博雯胡益丹郑晓诗
张帆林秋月*,郑博雯胡益丹郑晓诗
(1浙江师范大学,浙江省固体表面反应化学重点实验室,金华321004) (2浙江师范大学,化学与生命科学学院,金华321004)
含2-氨基吡啶及去甲基斑蝥酸根的钴(Ⅱ)配合物的晶体结构、与DNA和BSA相互作用及体外抗增殖活性
张帆1,2林秋月*,1,2郑博雯2胡益丹2郑晓诗2
(1浙江师范大学,浙江省固体表面反应化学重点实验室,金华321004) (2浙江师范大学,化学与生命科学学院,金华321004)
溶液法合成了二(去甲基斑蝥酸根)合钴(Ⅱ)酸二(2-氨基吡啶鎓)配合物(Hapy)2[Co(DCA)2]·6H2O(DCA=去甲基斑蝥酸根,C8H8O5;Hapy=2-氨基吡啶鎓,C5H7N2)。通过元素分析、摩尔电导、红外光谱、热重分析和X-射线单晶衍射对配合物进行了结构表征。该配合物为三斜晶系P1空间群,中心离子配位数为6。利用荧光光谱法和粘度法对配合物与DNA之间的相互作用进行了研究。结果表明,配合物以部分插入模式与DNA键合,猝灭常数Ksq为0.18。同时,利用荧光光谱研究了配合物与牛血清白蛋白(BSA)的相互作用。结果显示,配合物能与BSA发生较强的键合作用,键合常数Ka=3.88×106L·mol-1。体外抗增值活性结果显示,配合物对人肝癌细胞(SMMC-7721)和人胃癌细胞(MGC80-3)的抑制活性均强于去甲基斑蝥酸钠。
去甲基斑蝥酸根;2-氨基吡啶;钴(Ⅱ)配合物;与DNA和BSA相互作用
The interaction of transition metal complexes with biomacromolecules has received a great deal of attention during the past decade[1-2].Researchers find that the bioactivities of complexes could be improved by inserting drugsmolecules as ligands[3-4].Demethylcantharidin(NCTD,7-oxabicyclo[2,2,1]heptane-2,3-dicarbo-xylc acid anhydride),which is the derivative of cantharidin,has been used in clinical[5].Na2(DCA) (DCA=demethylcantharate)could inhibitprotein serine /threonine phosphatases(PP1,PP2A and PP2B)[6]. Meanwhile,themetal complexes containing demethylcantharate have been widely reported for their strong anti-tumour activities[7-8].Pyridine is an important class of N-containing heterocycles.The bioactivities of complexes containing pyridine ring have been reported,such as DNA binding property and antibacterial activity[9-10].The bioinorganic chemistry of cobalt has been rapidly expanded due to the increasing number of cobalt complexes of biological interest reported in the literature[11-12].
The complex(Hapy)2[Co(DCA)2]·6H2O was synthesized using cobalt acetate,NCTD and 2-aminopyridine(apy).The interaction of complex with DNA was investigated by fluorescence spectra and viscosity measurements.The interaction of complex with BSA was studied by fluorescence spectra.In addition,the antiproliferative activities of the complex against human hepatoma cells(SMMC-7721)and human gastric cancer cells(MGC80-3)were tested in vitro.
1 Experimental
1.1 M aterials and instruments
All reagents and chemicals were obtained from commercial sources.Demethylcantharidin(NCTD, C8H8O4)was obtained from Nanjing Zelang Medical Technology Co.Ltd.;Ethidium bromide(EB)was obtained from Fluka Co.;2-aminopyridine(apy, C5H6N2)and DNA was obtained from Sinopharm Chemical Reagent Co.Ltd.;DNA(ρ=200μg·mL-1, c=3.72×10-4mol·L-1),which A260/A280=1.8~2.0,was prepared by 50mmol·L-1NaCl;Bovine Serum Albumin (BSA)was purchased from Beijing BioDee BioTech Co.Ltd.and was stored at 277 K;BSA(ρ=500μg· mL-1,c=7.47×10-6mol·L-1)was prepared by 5 mmol· L-1NaCl;Human hepatoma cells(SMMC-7721)and human gastric cancer cells(MGC80-3)were purchased from Shanghai Institute of Cell Bank.Other chemical reagents in analytical reagent grade were used without further purification.
Elemental analyses of C,H and N were carried out in Vario ELⅢelemental analyzer.The molar conductance value was obtained from Orion 150Aplus conductometer.Infrared spectra were measured using the KBr disc method by NEXUS-670 FT-IR spectrometer in the spectral range 4 000~400 cm-1.The thermogravimetric analyses were monitored on TGA/ SDTA851ethermo gravimetric analyzer.Diffraction intensities of the complexes were collected at 293 K on Bruker SMART APEXⅡCCD difffractometer. Fluorescence emission spectra were obtained by Perkin-Elmer LS-55 spectrofluorometer.Viscosity experimentswere carried on Ubbelodhe viscometer.
1.2 Synthesis of the comp lex
The solution of Co(Ac)2·6H2O(0.5 mmol)and 2-aminopyridine(0.5 mmol)was stirred at the room temperature for 2 h.The solution of NCTD(0.5 mmol) was added dropwise in themixed solution.The pH of the solution was adjusted to 6.5 using dilute NaOH solution.The solution was then filtered after two hours.After two weeks,crystals with suitable size for single-crystal X-ray diffraction were obtained.
Anal.Calcd.for C26H42N4O6Co(%):C,43.00;H, 5.79;N,7.72.Found(%):C,42.81;H,5.97;N,7.68. IR spectra(KBr,cm-1):1 687(νas(COO-));1 413 (νs(COO-));1 266,10 29,985(ν(C-O-C)).The value of molar conductance of complex is 148 S·cm2·mol-1in 10-3mol·L-1DMF at 20℃,which suggest that the complex is 2∶1 type electrolyte[13].
1.3 DNA binding
1.3.1 Fluorescence spectra
Fluorescence quenching experiment was carried out by adding complex solution(0~2.50×10-4mol·L-1) to the samples containing 5.00×10-6mol·L-1EB and 7.44×10-5mol·L-1DNA.Themixture were diluted by Tris-HCl buffer solution(pH=7.4).Fluorescence was read at excitation wavelength(λex)of 252 nm andemissionwavelength(λem)between 500 nm and 700 nm.
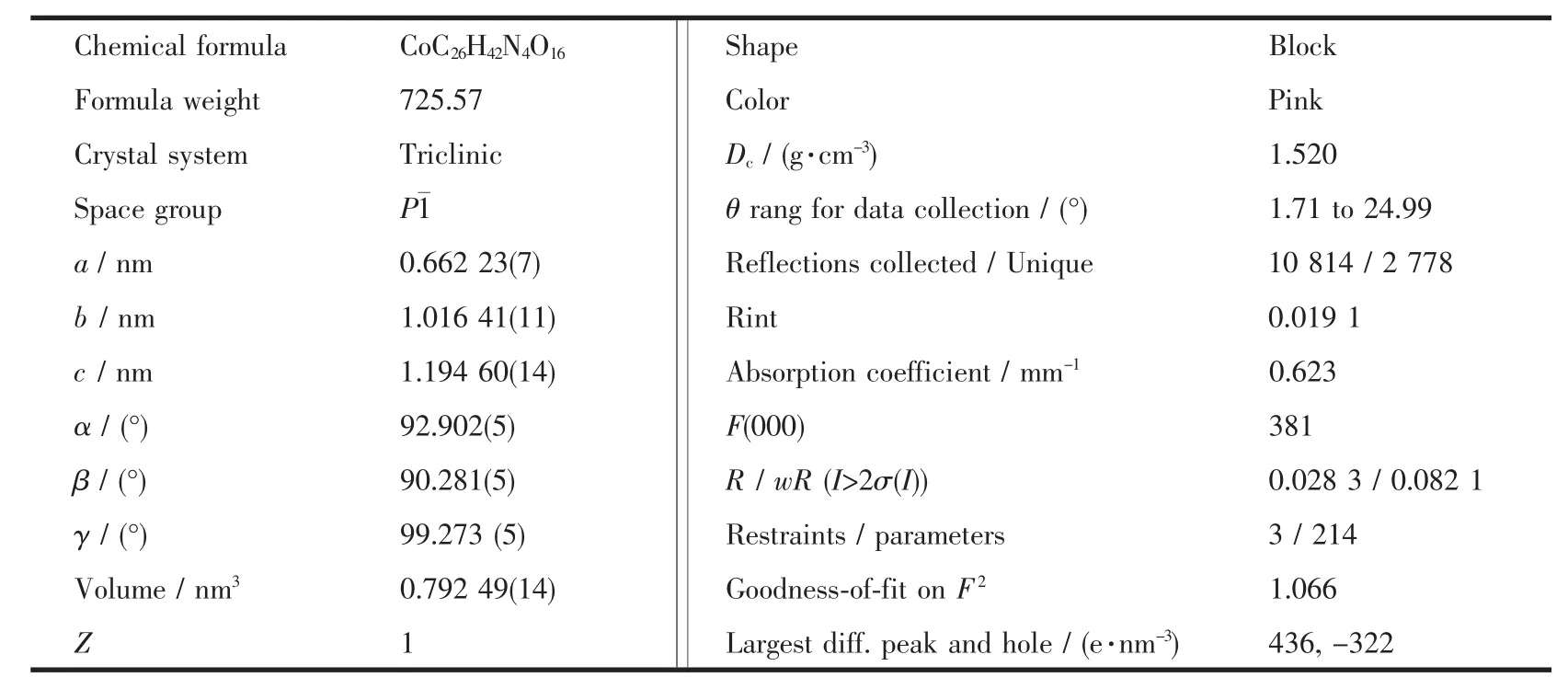
Table 1 Crystal data and structure refinement details for the complex
1.3.2 Viscosity measurements
Viscosity measurements were conducted on an Ubbelodhe viscometer.The compounds were added to DNA solution(3.72×10-4mol·L-1)by microsyringe. The concentration of the compounds were controlled in 0~3.33×10-6mol·L-1.The relative viscositiesη were calculated through equation[14]:η=(t-t0)/t0,where t0and t represent the flow time of DNA solution in the absence and presence of complex through the capillary,respectively.
1.4 Interaction w ith BSA
The complex(0~3.33×10-8mol·L-1)were added to themixed solution,which contains 4.98×10-7mol· L-1BSA and Tris-HCl buffer.Fluorescence quenching spectra were obtained by recording the emission spectra(255~500 nm)corresponding to excitation wavelength at255 nm.
1.5 Antiproliferative activity evaluation
The antiproliferative activities of Na2(DCA)and the complex were evaluated by human hepatoma cells (SMMC-7721)and human gastric cancer cells (MGC80-3).The antiproliferative activities were measured by the MTT assay.The compounds were dissolved in DMSO as 100 mmol·L-1stock solutions and diluted with culture medium before using.The final concentration of DMSO in the medium was less than 0.1%and it showed no interference with the biological activities tested[15].Cells were seeded for 24 h,and the complex or Na2(DCA)were added and incubated for 72 h.Then MTT(100 L,1 mg·mL-1, dissolved in culture medium)was added into each well and incubated for 4 h(37℃).The inhibition rate was calculated.The errors quoted were standard deviations,in which three replicates were used in the calculation[16].
1.6 Crystal structure determ ination
Single crystal,sized 0.167 mm×0.169 mm×0.246 mm,was analyzed by X-ray diffraction.The structure was solved by direct methods and refined by fullmatrix least-squares techniques using the SHELXTL-97 program package[17-18].All non-hydrogen atoms were refined anisotropically.Except the hydrogen atoms on oxygen atomswere located from the difference Fourier maps,the other hydrogen atoms were generated geometrically.Crystal data and experimental details for structural analyses are listed in Table 1.
CCDC:823705.
2 Results and discussion
2.1 Characterization and crystal structure
2.1.1 Thermogravimetric analysis(TGA)

Fig.1 TG-DTG curves of the complex
This experiment was performed in an air atmosphere with a heating rate of 10℃·min-1and temperature range of 30~800℃.The TG-DTG curves of complex are shown in Fig.1.According to the TGDTG curves,three steps of the weight loss processes were happened in this process.The first weight loss (14.03%)occurred in temperature range of 49~102℃, which corresponded to the departure of the six crystal water(14.90%theoretical).The second weight loss (25.61%),which corresponded to the departure of two 2-aminopyridinum(26.22%theoretical),was occurred in the temperature range of 103~251℃.The complex gave an acuity weight loss peak at temperature around 252~337℃,which corresponds to the weigh loss of 48.02%(48.56%theoretical).It indicates the thermal decomposition of two DCA.At temperature above 338℃,no further weight loss occurred.The sample residue was CoO,which weigh 11.67%of the initial mass(10.33%theoretical).
These results agree with the composition of the complex determined by elemental analyses and X-ray diffraction.
2.1.2 Structural description of the complex
The X-ray structural analysis shows that the complex crystallized in the triclinic crystal system with P1 space group.Molecular structure of the complex(Hapy)2[Co(DCA)2]·6H2O was showed in Fig. 2.Selected bond distances and angles were listed in Table 2.
The complex consisted cation part and anion part.Each Co(Ⅱ)ion was connected to two DCA ions, which formed anion part.The cation part was formed by two 2-aminopyridinum.Co(Ⅱ)ion was sixcoordinated by four carboxylate oxygen atoms O1,O3, O1A and O3A in four different carboxylate groups and two bridge oxygen atoms O5 and O5A from two demethylcantharates,which formed octahedral structure.Due to the binding of the bridge oxygen atoms O5 and O5A with Co(Ⅱ),four six-memberedrings(Co1-O1-C7-C2-C1-O5),(Co1-O3-C8-C3-C4-O5), (Co1-O1A-C7A-C2A-C1A-O5A)and(Co1-O3A-C8AC3A-C4A-O5A)were created.Two seven-membered rings(Co1-O1-C7-C2-C3-C8-O3)and(Co1-O1A-C7AC2A-C3A-C8A-O3A)were formed,which could have stabilized the anion partof the complex.
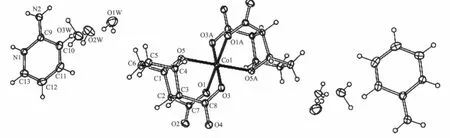
Fig.2 Labeled ORTEP diagram of the complex with 30%thermal probability ellipsoids shown

Table 2 Selected bond lengths(nm)and angles(°)for the com plex
2.2 DNA bingding studies
2.2.1 Fluorescence spectral studies
To study the mode and intensity of the interaction of complexes with DNA,EB was used as fluorescence probe.Fig.3 shows the emission spectra of EB bounded to DNA with 2-aminopyridine(Fig.3a) and the complex(Fig.3b)with similar peak shapes. Fig.3 suggested that,as the concentrations of the compounds increase,the emission intensity at 589 nm of EB-DNA system decrease in different degrees.
According to the Stern-Volmer equation[19]:F0/F= 1+Ksqr,where F0and F represented the fluorescence intensities in the absence or presence of complexes,r stands for the concentration ratio of the complexes to DNA.The quenching constant Ksqwas obtained as the slope of F0/F versus r linear plot,which were 0.058 (apy)and 0.18(complex).Results indicated that the complex could insert into the DNA base pairs,and release free EB from EB-DNA system.However, NCTD and Na2(DCA)can not quench the emission intensities of EB-DNA system significantly[20].
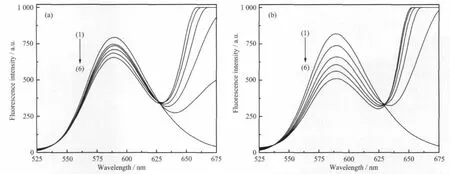
Fig.3 Fluorescence spectra of EB-DNA in the absence and presence of increasing the amountof compounds
2.2.2 Viscosity measurements

Fig.4 Effect of increasing amounts of the compounds on the relative viscosity of DNA at25℃
To further investigate the binding mode and binding intensity of the complexes with DNA,the DNA viscosity changing at 25℃was studied(Fig.4). The result showed that the relative viscosity of DNA didn′t change obviously after adding 2-aminopyridine and NCTD,while steadily decreased after adding complex,which suggested that the complex may partially insert into DNA[21].Structurally,the steric hindrance of the complex was accrescent due to the non-planar of DCA.Thus,the complex can only partially inserted into DNA.
2.3 Interaction w ith BSA
The fluorescence quenching of BSA by the complex was showed in Fig.5((a):complex;(b): NCTD).The results showed that BSA has strong fluorescence emission at 345 nm.The peak intensitydecreased with the increasing concentration of complex,which inferred that strong interactions and energy transfer between complex and BSA existed[22].

Fig.5 Fluorescence spectra of BSA in the absence and the presence of complex Inset:Stern-Volmer plots of the fluorescence titration data of the complexes
Assuming therewere n identical and independent binding sites in protein,the binding constant can be calculated using equation[23]:lg[(F0-F)/F]=lg Ka+n lg cQ, where F0and F represented the fluorescence intensities of BSA in the absence or presence of complex, cQstands for the concentration of the complex.Kaand n were calculated(see the slope and intercept of the line in illustration).The values of Kawere 2.78×104(NCTD)and 3.88×106L·mol-1(complex).The values of n were 0.66(NCTD)and 0.98(complex).The results suggested that strong binding force existed between the complex and the BSA with complex intensity stronger than NCTD.The binding sitewasone.
2.4 Antiproliferative activity evaluation
The antiproliferative activities of the Na2(DCA) and the complex against human hepatoma cells (SMMC-7721)(Fig.6)and human gastric cancer cells (MGC80-3)(Fig.7)were investigated in vitro.And the values of IC50for these compounds were listed in Table 3.The results showed complex had strong antiproliferative effect against two kinds of cells in the range of tested concentration.The inhibition effectwas enhanced by increasing the concentration of complex.

Data representmean+S.D.and all assayswere performed in triplicate for three independent experiments.*P<0.05,**P<0.01, ***P<0.001 vs Na2(DCA)in the same concentration,t-testFig.6 Inhibition effects of complex and Na2(DCA)on SMMC-7721 cell growth
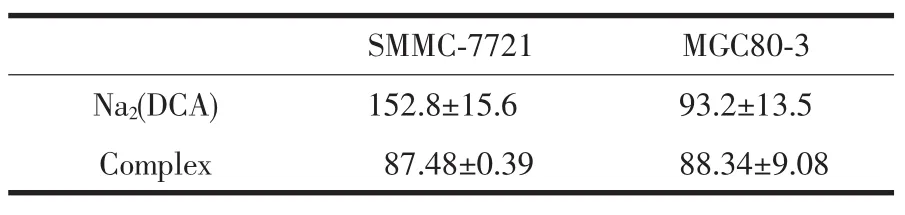
Table 3 IC50values(μmol·L-1)(72 h)of comp lex and Na2(DCA)on SMMC-7721 and MGC80-3 cells
As shown in Fig.6,the inhibition rates of complex were significantly higher than Na2(DCA)(P< 0.01)against human hepatoma cells.Especially at the concentration of 75.00μmol·L-1,the inhibition intensity of complex(52.2±1.9)was much stronger than Na2(DCA)(19.1±8.3).At concentration higher than 75.00μmol·L-1,the complex and Na2(DCA) exhibited similar efficiency.The values of IC50of complex((87.48±0.39)μmol·L-1)and Na2(DCA)((152.8 ±15.6)μmol·L-1)inferred that thecomplexhad strongeractivities against human hepatoma cells,even more intense than NCTD(IC50=(115.5±9.5)μmol·L-1)[24], which had been used in clinical.
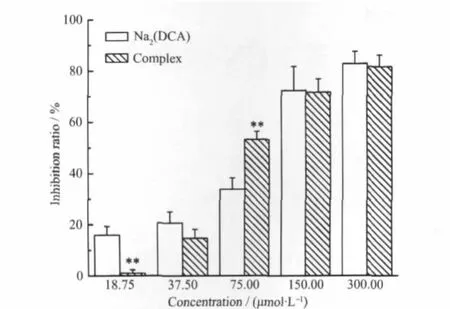
Data representmean+S.D.and all assays were performed in triplicate for three independent experiments.*P<0.05,**P<0.01, ***P<0.001 vs Na2(DCA)in the same concentration,t-testFig.7 Inhibition effects of complex and Na2(DCA)onMGC80-3 cell growth
As shown in Fig.7,the inhibition rate of complex were higher than the rate of Na2(DCA)(P<0.01) againsthuman gastric cancer cells at the concentration of 75.00μmol·L-1.At other tested concentrations,the inhibition ratesofcomplexwere close toNa2(DCA).
3 Conclusions
A novel cobalt(Ⅱ)complex was synthesized.The chemical formulaof the complexwas(Hapy)2[Co(DCA)2] ·6H2O.The structure of the complex was determined by X-ray diffraction.The complex crystallized in the triclinic crystal system with P1 space group.
All the tested results suggested that the complex could bind DNA via partial intercalation mode. Meanwhile,the complex could quench the fluorescence of BSA with the binding constant Kaof 3.88×106L·mol-1.
The antiproliferative activities testing revealed that the complex showed moderate inhibition ratios against human hepatoma cells and human gastric cancer cells in vitro.
Acknow ledgements:We thank Institute of Zhejiang Academy of Medical Sciences for helping with anti-proliferative activity test.Financial support,which is from Natural Science Foundation of Zhejiang Province,China(Grant No.Y407301),is gratefully acknowledged.
[1]ZHOU Jian-Liang(周建良),CHUN Xiao-Gai(春晓改), ZHOU Lin-Jiao(周琳姣),et al.Chinese J.Inorg.Chem. (WujiHuaxue Xuebao),2010,26(4):645-650
[2]Wu SS,Yuan W B,Wang H Y,et al.J.Inorg.Biochem., 2008,102(11):2026-2034
[3]Skyrianou K C,Perdih F,Turel I,et al.J.Inorg.Biochem., 2010,104(7):740-749
[4]Buchtík R,Trávnícˇ ek Z,Van cˇ o J,etal.Dalton Trans.,2011, 40(37):9404-9412
[5]Yin F L,Zou J J,Xu L,et al.J.Rare Earths,2005,23(5): 596-599
[6]Hill T A,Stewart SG,Gordon C P,et al.ChemMedChem, 2008,3(12):1878-1892
[7]Wang Y Y,Hu R D,Lin Q Y,et al.Asian J.Chem.,2010, 22(8):5993-5999
[8]Lin QY,Wang Y Y,Feng Y L,etal.J.Coord.Chem.,2011, 64(5):920-930
[9]Bhat S S,Kumbhar A S,Lnnecke P,et al.Inorg.Chem., 2010,49(11):4843-4853
[10]Abu-Youssef M A M,Soliman SM,Langer V,et al.Inorg. Chem.,2010,49(21):9788-9797
[11]Peng B,Chao H,Sun B,et al.J.Inorg.Biochem.,2007, 101(3):404-411
[12]ZHAO Guo-Liang(赵国良),SHIXia(施霞),ZHANG Jun-Ping(张均平),et al.Sci.China Chem.(Zhongguo Kexue, Huaxue),2010,40(10):1525-1535
[13]Geary W J.Coord.Chem.Rev.,1971,7(1):81-122
[14]Nagababu P,Satyanarayana S.Polyhedron,2007,26(8):1686 -1692
[15]Kumar C SA,Prasad SB B,Vinaya K,et al.Eur.J.Med. Chem.,2009,44(2):834-844
[16]Zheng X L,Sun H X,Liu X L,et al.Acta Pharmacol.Sin., 2004,25(8):1090-1095
[17]Sheldrick G M.SHELXS-97,Program for the Solution of Crystal Structures,University of Göttingen,Germany,1997.
[18]Sheldrick GM.SHELXL-97,Program for the Refinement of Crystal Structures,University of Göttingen,Germany,1997.
[19]LIN Qing-Bin(林庆斌),ZENG Xi-Rui(曾锡瑞),NIChun-Lin(倪春林),et al.Chinese J.Inorg.Chem.(Wuji Huaxue Xuebao),2007,23(8):1341-1346
[20]Zhu W Z,Lin Q Y,Lu M,et al.J.Fluoresc.,2009,19(5): 857-866
[21]Bhat I H,Tabassum S.Spectrochim.Acta A,2009,72(5): 1026-1033
[22]Wang Y J,Hu R D,Jiang D H,et al.J.Fluoresc.,2011,21 (2):813-823
[23]Patra A,Sarkar S,Mukherjee T,et al.Polyhedron,2011,30 (17):2783-2789
[24]Wang N,Lin Q Y,Feng J,et al.Inorg.Chim.Acta,2010, 363(13):3399-3406
Crystal Structure,Interaction w ith DNA and BSA and Antiproliferative Activities of Cobalt(Ⅱ)Com p lex w ith Demethylcantharate and 2-Am inopyridine
ZHANG Fan1,2LIN Qiu-Yue*,1,2ZHENG Bo-Wen2HU Yi-Dan2ZHENG Xiao-Shi2
(1Zhejiang Key Laboratory for Reactive Chemistry on Solid Surfaces,Zhejiang Normal University,Jinhua,Zhejiang 321004,China)
(2College of Chemical and Life Science,Zhejiang Normal University,Jinhua,Zhejiang 321004,China)
A cobalt(Ⅱ)complex(Hapy)2[Co(DCA)2]·6H2O(DCA=demethylcantharate,7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylate,C8H8O5;Hapy=2-aminopyridinum,C5H7N2)was synthesized and characterized by elemental analysis,molar conductance,infrared spectra,thermogravimetric analysis and X-ray diffraction.The complex crystallized in the triclinic crystalsystem,P1 space group,with a=0.662 23(7)nm,b=1.016 41(11)nm,c=1.194 60(12) nm,V=0.792 49(14)nm3,Z=1,Dc=1.520 g·cm-3,Mr=725.57,λ(Mo Kα)=0.071 073 nm,F(000)=381,R=0.028 3 and wR=0.082 1(I>2σ(I)).The coordination number ofmetal ion was six and DCA ionswere tridentate ligand.DNA binding property of the complex was investigated by fluorescence spectra and viscosity measurements.Results indicated the complex could bind to DNA via partial intercalationmode.The value of quenching constant Ksqwas 0.18.The interaction of the complex with bovine serum albumin(BSA)was also studied by fluorescence spectra. The results suggested that the complex could quench the fluorescence of BSA with the binding constant Kaof 3.88×106L·mol-1.The antiproliferative activity test revealed that complex showed stronger inhibition ratios against human hepatoma cells(SMMC-7721)lines and human gastric cancer cells(MGC80-3)lines than Na2(DCA)in vitro.CCDC:823705.
demethylcantharate;2-aminopyridine;cobalt(Ⅱ)complex;interaction with DNA and BSA
book=0,ebook=53
O614.81+2
A
1001-4861(2012)11-2451-07
2012-01-11。收修改稿日期:2012-06-26。浙江省自然科学基金(No.Y407301)资助项目。
*通讯联系人。E-mail:sky51@zjnu.cn
