Induction, modulation and potential targets of miR-210 in pancreatic cancer cells
2012-07-07
Beijing, China
Induction, modulation and potential targets of miR-210 in pancreatic cancer cells
Wei-Yun Chen, Wen-Jing Liu, Yu-Pei Zhao, Li Zhou, Tai-Ping Zhang, Ge Chen and Hong Shu
Beijing, China
BACKGROUND: MiR-210 is induced by hypoxia and plays different roles in the development of certain cancers. However, little is known about its role in pancreatic cancer (PC). This study aimed to explore the induction and modulation of PC by miR-210 and its potential molecular targets.
METHODS: PC cells were cultured under normoxic and hypoxic conditions. Expression of miR-210 and hypoxia-inducible factor (HIF)-1α was detected using quantitative reverse-transcription polymerase chain reaction. Cancer cells were transiently transfected with HIF-1α small interfering RNA (siRNA) and miR-210 mimics, and cell proliferation was measured using the CCK-8 assay. Potential targets for miR-210 were then identified using a dual luciferase reporter assay.
RESULTS:Hypoxic conditions induced miR-210 expression in six PC cell lines (AsPC-1, BxPC-3, MIAPaCa-2, PANC-1, Su86.86 and SW1990), but not in Capan-1 or T3M4 cells. Transfection of HIF-1α siRNA into PANC-1 cells markedly inhibited HIF-1α expression, and subsequently downregulated miR-210 expression under hypoxic conditions. MiR-210 had no observable impact on the proliferation of PANC-1 or Su86.86 cells and dual luciferase reporter assays showed significantly reduced luciferase activity in the wildtype E2F3, EFNA3, GIT2, MNT, ZNF462 and EGR3 constructs, compared to the corresponding mutants, but not in HOXA3. CONCLUSIONS: These results suggest that miR-210 expression in PC cells is induced by hypoxia through a HIF-1α-dependent pathway, but does not influence PC cell proliferation. Also, E2F3, EFNA3, GIT2, MNT, ZNF462 and EGR3 may be potential miR-210 targets in PC.
(Hepatobiliary Pancreat Dis Int 2012;11:319-324)
miR-210; hypoxia; HIF-1α; pancreatic cancer; proliferation
Introduction
Pancreatic cancer (PC) is a lethal solid malignant tumor with an extremely dismal prognosis.[1]The estimated mortality is almost the same as the estimated incidence, with an overall 5-year survival rate of less than 5%.[2]Therefore, the molecular pathogenesis of PC has been the subject of much research to explore new therapeutic targets and improve treatment efficacy.[1]K-ras, CDKN2A, P53 and DPC4 are known to be mutated or inactivated during PC tumorigenesis, and they mediate other functional and relevant cancer pathways.[3-5]However, further studies are needed.
MicroRNAs (miRs: highly conserved, non-coding, 17-25 nt RNAs) are thought to modulate gene expression at the post-transcriptional level.[6,7]MiRs are also associated with the main phenotypes of many cancer cells (including PC), such as proliferation, invasion and apoptosis.[8-10]MiR-210 is differentially expressed in different neoplasms,[11-16]and cell-line-based functional studies have provided controversial results.[17-19]Therefore, the effects of miR-210 may be cell-type specific. Few studies of PC have focused on the induction, modulation, targeting, and clinical and prognostic significance of miR-210,[19-21]and controversies remain. For example, miR-210 shows antitumor activity in a human PC-bearing nude mouse model, but acts as an independent marker for poor prognosis in patients with PC.[19,20]Therefore, the present study aimed to address these issues using several PC cell lines.
Methods
Cell culture and hypoxic conditions
Eight human pancreatic cancer cell lines were provided by Professor Helmut Freiss, Heidelberg University,Germany. AsPC-1, BxPC-1, Su86.86 and SW1990 were cultured in Dulbecco's modified Eagle's medium (DMEM, Hyclone, Thermo Fisher Scientific Inc., Waltham, MA) supplemented with 10% fetal bovine serum (FBS, Hyclone). Capan-1, MIAPaCa-2, PANC-1 and T3M4 were cultured in RPMI-1640 (Hyclone) supplemented with 10% FBS. All cells were cultured in a humidified incubator with 5% CO2at 37 ℃. For the hypoxic experiments, each cell line was split into two identical plates 24 hours before treatment: one was placed in an airtight chamber flushed with a gas mixture of 93% N2, 5% CO2, and 2% O2, while the other was kept under normoxic conditions. The same temperature was maintained for all treatment groups by placing the sealed chambers inside the incubator used for normoxic conditions.
Transient transfection
PANC-1 and Su86.86 cells, in which miR-210 is differentially expressed and induced,[19]were transiently transfected with small interfering RNA (siRNA) targeting hypoxia-inducible factor (HIF)-1α or a scrambled sequence (described in a previous article[22]and shown in Table 1), and miR-210 mimics as well as negative control (Gene-Pharma), using lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
Quantitative reverse-transcription polymerasechain reaction (qRT-PCR)
Total RNA was extracted using TRIzol (Invitrogen). Hsa-miR-210 and internal control RNU6B TaqMan qRT-PCR kits were from Applied Biosystems. Reverse transcription was performed using the PrimeScript RT reagent kit (Takara) according to the manufacturer's instructions after total RNA extraction. The PCR conditions were: pre-denaturation step at 95 ℃ for 30 seconds followed by 40 cycles of 95 ℃ for 5 seconds and 60 ℃ for 30 seconds, and then another 95 ℃ for 60 seconds and 55 ℃ for 60 seconds. GAPDH was used as an internal control. The primer sequences are shown in Table 2.
Prediction of miR-210 targets
Bioinformatic prediction of miR-210 target genes was performed using three programs: TargetScan (Ver. 5.1), PicTar (Ver. 2006), and MiRanda (Ver. 2008). Common targets identified by at least two of the programs, and genes likely to be related to tumor growth regulation were considered for further experimental analysis.
Cell proliferation assay
Cell proliferation was determined using the cellcount kit, CCK-8. Briefly, cells were seeded in 96-well plates at 5×103cells/well and transfected with various concentrations of miR-210 mimics or a negative control. The CCK-8 assay was performed after 48 hours. After 4 hours of incubation with culture medium containing the CCK-8 reagent, the absorbance was read at 450 nm using a microplate enzyme-linked immunosorbent assay reader (Labsystems Dragon, Wellscan).
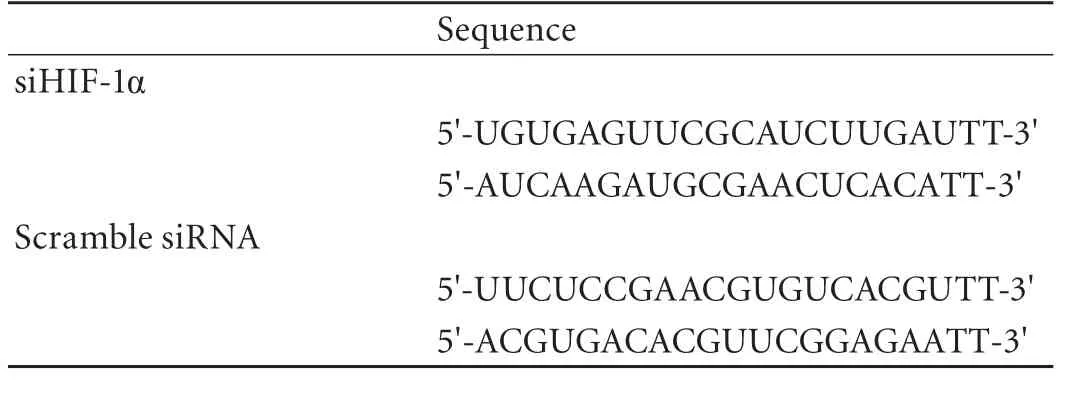
Table 1. Sequences used for siRNA targeting HIF-1α

Table 2. Primer sequences used for qRT-PCR
Dual-luciferase reporter assay
First, fragments containing the putative miR-210 target site and the immediately surrounding sequences within the 3'UTR of the candidate target genes were constructed. The fragments were subcloned into the pRL-TK vector (Promega) downstream of theRenillaluciferase gene. The fragments in which the sequence complementary to miR-210 was mutated were used as controls, and subcloned into the pRL-TK vector. The sequences used to construct the luciferase reporter are shown in Table 3. In the experimental group, theRenillaluciferase reporter vector containing a putative miR-210 target site (or control sequence) was co-transfected into PANC-1 cells along with miR-210 mimics (Gene-Pharma) using lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The empty firefly luciferase vector, pGL3, was co-transfected to serve as a transfection normalization reporter. Luciferase activity was measured using a Dual-luciferase Reporter Assay System (Promega) and a Modulus Microplate Multimode Reader (Turner BioSystems).
Statistical analysis
Differences between continuous variables werecalculated using Student'sttest (independent samples). APvalue of <0.05 was considered statistically significant.

Table 3. Sequences used for construction ofRenillaluciferase reporter vectors
Results
Expression of miR-210 in PC cell lines undernormoxic and hypoxic conditions
The qRT-PCR data revealed that miR-210 expression was induced by hypoxia in six of the PC cell lines: AsPC-1, BxPC-3, MIAPaCa-2, PANC-1, Su86.86 and SW1990 (P<0.05, Fig. 1), but not in Capan-1 and T3M4 cells (P>0.05, Fig. 1).
HIF-1α-dependent miR-210 induction by hypoxia
To investigate the mechanisms underlying miR-210 induction in PC cell lines under hypoxic conditions, an siRNA targeting HIF-1α was constructed and transfected into hypoxic PANC-1 cells. HIF-1α expression was significantly reduced in cells transfected with siHIF-1α after 24 hours compared with that in control cells transfected with scrambled siRNA instead of siHIF-1α (Fig. 2A). qRT-PCR showed that miR-210 expression was lower in cells transfected with siHIF-1α than in controlcells under hypoxic conditions (P<0.01, Fig. 2B), but no difference was found under normoxic conditions (P>0.05, Fig. 2B).
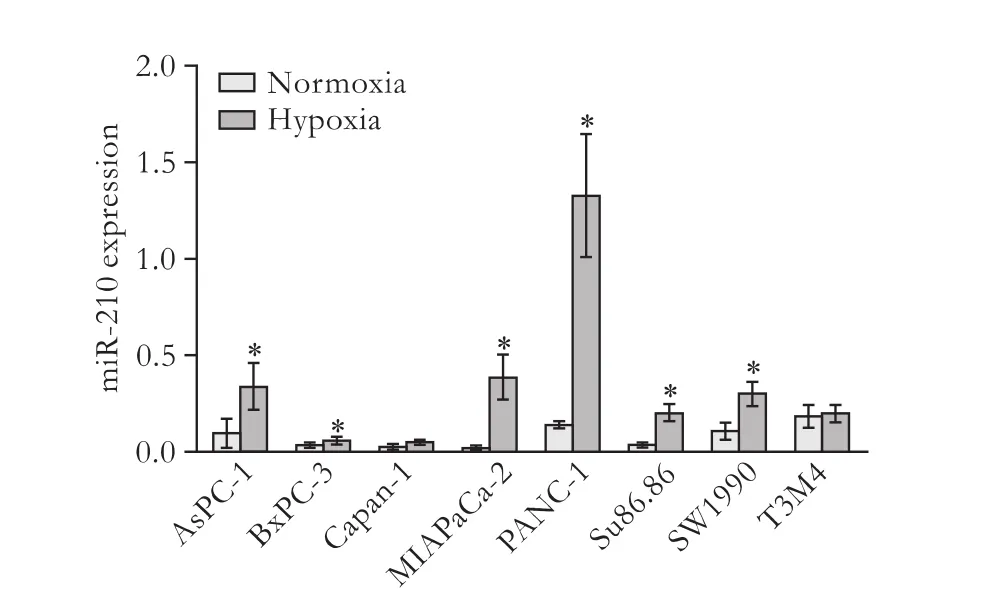
Fig. 1. MiR-210 expression is broadly induced by hypoxia in PC cell lines. Up-regulation of miR-210 by hypoxia was detected in 6/8 PC cell lines: AsPC-1, BxPC-3, MIAPaCa-2, PANC-1, Su86.86 and SW1990. Data represent the mean±SD of three independent experiments. *:P<0.05.
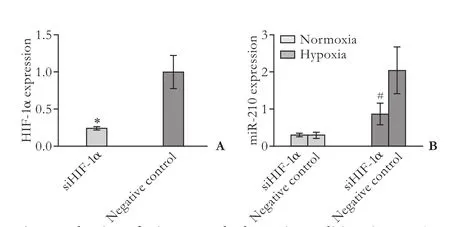
Fig. 2. Induction of miR-210 under hypoxic conditions in PANC-1 cells is HIF-1α-dependent. A: siRNA targeted to HIF-1α reduced the expression of HIF-1α in PANC-1 cells; B: siRNA targeted to HIF-1α reduced the expression of miR-210 in PANC-1 cells under hypoxic conditions, but not under normoxic conditions. Data represent the mean±SD of three independent experiments. *:P<0.05; #:P<0.01.
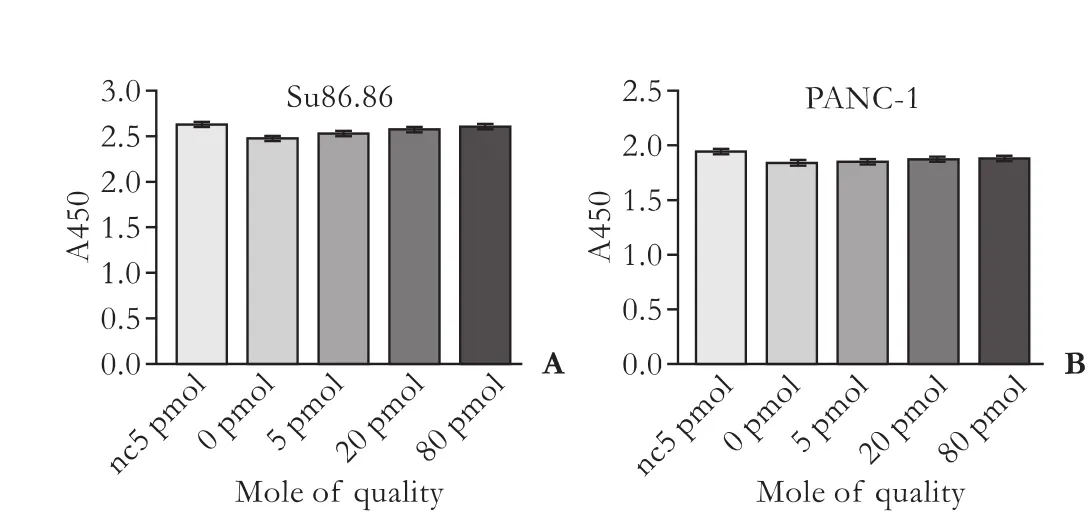
Fig. 3. Impact of miR-210 on the proliferation of two PC cell lines measured using the CCK-8 assay. A: Su86.86 cells; B: PANC-1 cells. Data represent the mean±SD of three independent experiments.
Effect of miR-210 on PANC-1 and Su86.86 PC cell proliferation
MiR-210 had no effect on the proliferation of PANC-1 or Su86.86 cells (P>0.05, Fig. 3).
Identification of potential targets of miR-210 in PANC-1 cells
TargetScan, PicTar and MiRanda identified E2F3, EFNA3, EGR3, GIT2, HOXA3, MNT and ZNF462 as putative targets for miR-210. A dual-luciferase reporter assay showed a reduction of luciferase activity in the wildtype E2F3 (P<0.0001), EFNA3, GIT2, MNT, ZNF462 (P<0.005) and EGR3 constructs (P<0.05) compared with the corresponding control (mutant) constructs. However, there was no difference in luciferase activity between the wild-type and mutant HOXA3 constructs, indicating that E2F3, EFNA3, EGR3, GIT2, MNT and ZNF462 are potential targets for miR-210 in PANC-1 cells (Fig. 4).
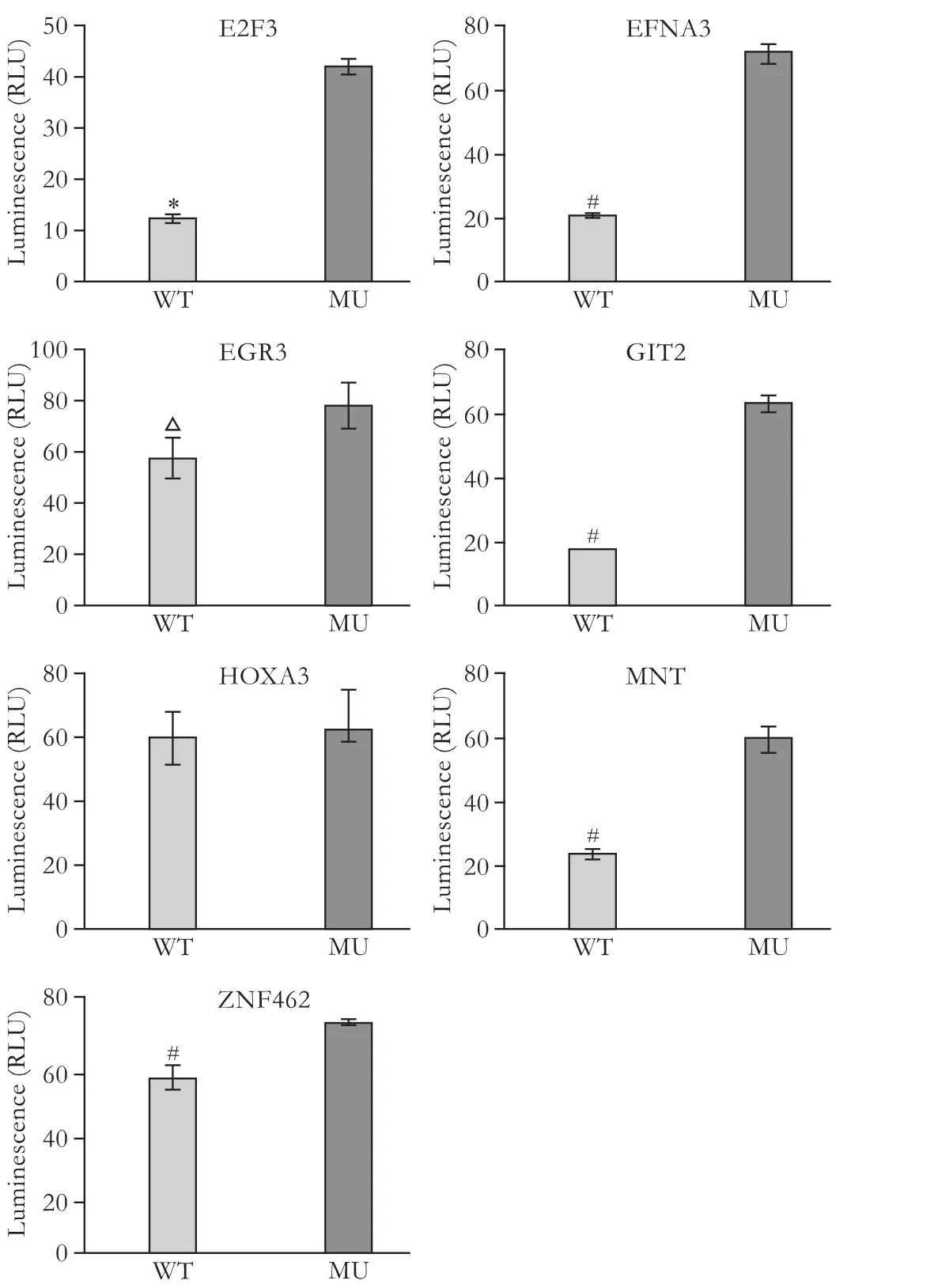
Fig. 4. Identification of potential miR-210 targets in PANC-1 cells using the dual-luciferase reporter assay. A significant negative effect on luciferase activity was found for the wild-type E2F3, EFNA3, GIT2, MNT, ZNF462 and EGR3 constructs compared with the corresponding mutant constructs (but not HOXA3) in PANC-1 cells. Data represent the mean±SD of three independent experiments. *:P<0.0001; #:P<0.005; △:P<0.05.
Discussion
Hypoxia mediates cancer cell resistance to treatment, is associated with a poor prognosis, drives genomic instability, and alters DNA damage repair pathways.[23]HIF-1α plays a key role in many important pathological process.[24]Recently, miRNA, non-coding small RNA, was found to be involved in the induction of certain cancer cell phenotypes.[25]MiR-210 is induced by hypoxia in breast cancer cells through an HIF-1αdependent pathway.[26,27]However, there is only a single study reporting miR-210 induction in hypoxic PC cells.[19]The present study confirmed the findings of this study in a majority of PC cell lines, indicating thathypoxia-induced miR-210 expression is a common event in the development of PC. However, this was not the case in two cell lines, Capan-1 and T3M4. The data also indicated that miR-210 expression in these cell lines needs further verification. In addition, siRNA transfection blocked HIF-1α and effectively downregulated the expression of HIF-1α and miR-210. Thus, HIF-1α may be a crucial modulator of miR-210 expression in PC cells, as previously reported for RCC4/VHL cells.[19]The results of the present study also indicated that the HIF-1α-dependent pathway is involved in regulating miR-210 expression in different malignant cell types; thus, the possibility that related molecules may serve as novel therapeutic targets cannot be ignored.
Because evidence shows that miR-210 expression is associated with poor PC prognosis,[20]we hypothesized that miR-210 promotes PC cell growth. Surprisingly, a previous study showed that miR-210 has no effect on the proliferation of Su86.86 cells; indeed, it actually inhibits tumor growth in a xenograft model.[19]These data are inconsistent with previous clinical studies.[20]Therefore, we attempted to evaluate the impact of miR-210 on PC cell proliferation. Our results also showed that miR-210 transfection had no effect on the proliferation of Su86.86 or PANC-1 cells. This suggests that miR-210 has neither a positive nor a negative effect on PC cell growthin vitro. Therefore, thein vivoeffects of the miRNA cannot be explained by regulation of PC cell proliferation, and a novel mechanism should be proposed.
Although some genes have been identified as miR-210 targets in several cancers,[11,17-19,23]none of these studies investigated PC. The present study is the first to identify E2F3, EFNA3, GIT2, MNT, ZNF462 and EGR3 as potential targets for miR-210 in PC using a dual-luciferase reporter assay. The genes were identified by the computational prediction programs, TargetScan, PicTar and MiRanda. Of the candidate genes, E2F3, MNT and EFNA3 have been identified as targets for miR-210 in ovarian, colon and breast cancer cells.[11,17,19]The fact that MNT and E2F3 inhibit and promote cancer cell proliferation suggest that a balance between antiproliferative and pro-proliferative events might account, at least in part, for the observed effects of miR-210 on PC cells.[17,28]More importantly, this is the first report showing that GIT2, ZNF462 and EGR3 are potential miR-210 targets in PC. Thus far, a few reports have linked these genes to cancer,[29,30]but none are related to miRNA. Therefore, further studies on the relationship between these genes and miRNA in cancer are expected. Unlike miR-10a,[31]the present study did not identify HOXA3 as a putative miR-210 target. Furthermore, additional mechanistic studies should be focused on the role and regulation of novel targets in PC.
In conclusion, the results of the present study showed that miR-210 is largely induced by hypoxia via an HIF-1α-dependent pathway in PC cells, but does not affect cell proliferation. Also, E2F3, EFNA3, GIT2, MNT, ZNF462 and EGR3 are potential miR-210 targets in PC.
Acknowledgements: We thank Dr. Chun-Jing Bian for his direction on experimental techniques.
Contributors: CWY and LWJ contributed equally to this work. CWY, LWJ and ZYP proposed the study. CWY and LWJ wrote the first draft. CWY, LWJ and ZL analyzed the data. All authors contributed to the design and interpretation of the study and to further drafts. ZYP is the guarantor.
Funding: This study was supported by grants from the National Laboratory of Molecular Biology Special Foundation (2060204) and the Beijing Municipal Natural Science Foundation (7100003), China. Ethical approval: Not needed.
Competing interest: No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
1 Hidalgo M. Pancreatic cancer. N Engl J Med 2010;362:1605-1617.
2 Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA Cancer J Clin 2008;58:71-96.
3 Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005; 7:469-483.
4 Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci U S A 2006;103:5947-5952.
5 Feldmann G, Beaty R, Hruban RH, Maitra A. Molecular genetics of pancreatic intraepithelial neoplasia. J Hepatobiliary Pancreat Surg 2007;14:224-232.
6 Ambros V. The functions of animal microRNAs. Nature 2004;431:350-355.
7 Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-297.
8 Zaman MS, Chen Y, Deng G, Shahryari V, Suh SO, Saini S, et al. The functional significance of microRNA-145 in prostate cancer. Br J Cancer 2010;103:256-264.
9 Yu S, Lu Z, Liu C, Meng Y, Ma Y, Zhao W, et al. miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer. Cancer Res 2010;70:6015-6025.
10 Schickel R, Park SM, Murmann AE, Peter ME. miR-200c regulates induction of apoptosis through CD95 by targeting FAP-1. Mol Cell 2010;38:908-915.
11 Giannakakis A, Sandaltzopoulos R, Greshock J, Liang S, Huang J, Hasegawa K, et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther 2008;7:255-264.
12 Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, et al. Detection of elevated levels of tumourassociated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol 2008;141:672-675.
13 Cho WC, Chow AS, Au JS. Restoration of tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung adenocarcinoma patients with epidermal growth factor receptor mutation. Eur J Cancer 2009;45:2197-2206.
14 Tömböl Z, Szabó PM, Molnár V, Wiener Z, Tölgyesi G, Horányi J, et al. Integrative molecular bioinformatics study of human adrenocortical tumors: microRNA, tissue-specific target prediction, and pathway analysis. Endocr Relat Cancer 2009;16:895-906.
15 Satzger I, Mattern A, Kuettler U, Weinspach D, Voelker B, Kapp A, et al. MicroRNA-15b represents an independent prognostic parameter and is correlated with tumor cell proliferation and apoptosis in malignant melanoma. Int J Cancer 2010;126:2553-2562.
16 Miko E, Czimmerer Z, Csánky E, Boros G, Buslig J, Dezso B, et al. Differentially expressed microRNAs in small cell lung cancer. Exp Lung Res 2009;35:646-664.
17 Zhang Z, Sun H, Dai H, Walsh RM, Imakura M, Schelter J, et al. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle 2009; 8:2756-2768.
18 Favaro E, Ramachandran A, McCormick R, Gee H, Blancher C, Crosby M, et al. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS One 2010;5:e10345.
19 Huang X, Ding L, Bennewith KL, Tong RT, Welford SM, Ang KK, et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell 2009; 35:856-867.
20 Greither T, Grochola LF, Udelnow A, Lautenschläger C, Würl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer 2010;126:73-80.
21 Ho AS, Huang X, Cao H, Christman-Skieller C, Bennewith K, Le QT, et al. Circulating miR-210 as a novel hypoxia marker in pancreatic cancer. Transl Oncol 2010;3:109-113.
22 Li J, Shi M, Cao Y, Yuan W, Pang T, Li B, et al. Knockdown of hypoxia-inducible factor-1alpha in breast carcinoma MCF-7 cells results in reduced tumor growth and increased sensitivity to methotrexate. Biochem Biophys Res Commun 2006;342:1341-1351.
23 Bristow RG, Hill RP. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat Rev Cancer 2008;8: 180-192.
24 Wilkinson S, Ryan KM. Growth factor signaling permits hypoxia-induced autophagy by a HIF1alpha-dependent, BNIP3/3L-independent transcriptional program in human cancer cells. Autophagy 2009;5:1068-1069.
25 Loayza-Puch F, Yoshida Y, Matsuzaki T, Takahashi C, Kitayama H, Noda M. Hypoxia and RAS-signaling pathways converge on, and cooperatively downregulate, the RECK tumor-suppressor protein through microRNAs. Oncogene 2010;29:2638-2648.
26 Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, et al. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res 2008;14:1340-1348.
27 Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res 2009;69:1221-1229.
28 Olsson AY, Feber A, Edwards S, Te Poele R, Giddings I, Merson S, et al. Role of E2F3 expression in modulating cellular proliferation rate in human bladder and prostate cancer cells. Oncogene 2007;26:1028-1037.
29 Sabe H, Onodera Y, Mazaki Y, Hashimoto S. ArfGAP family proteins in cell adhesion, migration and tumor invasion. Curr Opin Cell Biol 2006;18:558-564.
30 Kokkinakis DM, Brickner AG, Kirkwood JM, Liu X, Goldwasser JE, Kastrama A, et al. Mitotic arrest, apoptosis, and sensitization to chemotherapy of melanomas by methionine deprivation stress. Mol Cancer Res 2006;4:575-589.
31 Han L, Witmer PD, Casey E, Valle D, Sukumar S. DNA methylation regulates MicroRNA expression. Cancer Biol Ther 2007;6:1284-1288.
July 15, 2011
Accepted after revision October 20, 2011
Author Affiliations: Department of General Surgery, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences/Peking Union Medical College, Beijing 100730, China (Chen WY, Liu WJ, Zhao YP, Zhou L, Zhang TP, Chen G and Shu H)
Yu-Pei Zhao, MD, Department of General Surgery, Peking Union Medical College Hospital, National Laboratory of Medical Molecular Biology, Chinese Academy of Medical Sciences/Peking Union Medical College, Beijing 100730, China (Tel: 86-10-65296007; Fax: 86-10-65124875; Email: zhao8028@263.net)
© 2012, Hepatobiliary Pancreat Dis Int. All rights reserved.
10.1016/S1499-3872(12)60168-4
杂志排行
Hepatobiliary & Pancreatic Diseases International的其它文章
- Management of splenic artery aneurysm associated with extrahepatic portal vein obstruction
- Pancreas-preserving segmental duodenectomy for gastrointestinal stromal tumor of the duodenum and splenectomy for splenic angiosarcoma
- Quantitative analysis of intestinal gas in patients with acute pancreatitis
- Can the biliary enhancement of Gd-EOB-DTPA predict the degree of liver function?
- Quality control measures for lowering the seroconversion rate of hemodialysis patients with hepatitis B or C virus
- Effect of recombinant human growth hormone and interferon gamma on hepatic collagen synthesis and proliferation of hepatic stellate cells in cirrhotic rats
