Expression of SOCS3 throughout liver regeneration is not regulated by DNA methylation
2012-07-07GangGuoJieXiaJiBaoHuaiQiangSunYuJunShiandHongBu
Gang Guo, Jie Xia, Ji Bao, Huai-Qiang Sun, Yu-Jun Shi and Hong Bu
Chengdu, China
Expression of SOCS3 throughout liver regeneration is not regulated by DNA methylation
Gang Guo, Jie Xia, Ji Bao, Huai-Qiang Sun, Yu-Jun Shi and Hong Bu
Chengdu, China
BACKGROUND:While suppressor of cytokine signaling 3 (SOCS3) plays a crucial role in suppressing dysplasia and tumorigenesis, it also offers a typical instance of DNA methylation in the regulation of gene transcription, since the promoter region of the SOCS3 gene is rich in CpG islands (CGIs). During liver regeneration initiated by partial hepatectomy, SOCS3 acts as a suppressor to balance the acute-phase response and terminate the regeneration. This study aimed to determine whether the variation of SOCS3 expression throughout liver regeneration is also regulated by its DNA methylation.
METHODS:We established a 70% partial hepatectomy mouse model and the animals were sacrificed at indicated times to assess the SOCS3 expression. We performed bisulfite sequencing PCR and DNA sequencing to investigate the detailed cytosine methylation in the SOCS3 gene.
RESULTS:Within the promoter sequence, 58 CGIs were identified and 30 were found variously methylated before or after operation; however, methylation remained at a very low level. No evidence indicated that the total methylation level or the methylation of any CpG site regularly changed throughout liver regeneration.
CONCLUSION:DNA methylation or demethylation seems to be a relatively stable modification of cytosine, but not a dynamic and reversible process to regulate gene transcription in daily and acute pathophysiological events.
(Hepatobiliary Pancreat Dis Int 2012;11:401-406)
liver regeneration; suppressor of cytokine signaling 3; DNA methylation
Introduction
Liver regeneration after partial hepatectomy (PH) is a unique growth process in which the hepatic mass is rapidly reconstituted after surgical removal of two-thirds of the liver. The regenerative process after PH is dependent on the replication of mature hepatocytes, which are normally quiescent.[1-3]A large number of cytokines,[4-7]including IL-6 and tumor necrosis factor (TNF), and growth factors, including hepatocyte growth factor (HGF) and epithelial growth factor (EGF), participate in the initiation of liver regeneration. As one of the main mediators of the acute-phase response, IL-6 is a critical component of a cytokine pathway activated very shortly after PH that involves signaling through TNFR1 and activation of the transcription factor NF-κB and the signal transducers and activators of transcription (STATs).[8]STATs are activated through phosphorylation by Janus kinases (JAKs), and the activated STATs then dimerize and bind to DNA to activate the transcription of target genes.[9]Suppressor of cytokine signaling (SOCS) genes are the targets of STATs, and encode eight proteins that inhibit a variety of signaling pathways.[10]SOCS proteins appear to inhibit cytokine signaling by targeting different components of the signaling complex either (a) by directly binding to activated, tyrosine-phosphorylated cytokine receptors or JAKs, or (b) by targeting receptor complexes for proteosomal degradation. Thus, stimulation of SOCS transcription by STATs establishes a negative feedback loop that inhibits the ongoing activation of cytokine signaling.[11-13]In the liver, SOCS3 is a critical inhibitor of IL-6 and STATs signaling and has a major effect on the acute-phase response to liver injury or inflammation.[14]Previous data showed that SOCS3 is highly expressed between 2 and 8 hours after PH, which partially overlaps with the time-frame of STAT3 expression.[15]
DNA methylation is the most studied epigenetic mechanism and CpG methylation is central to many biological processes and human diseases.[16,17]Aberrant hypermethylation of CpG islands (CGIs) in promoter regions is associated with the transcriptionalsuppression of various genes in several types of epithelial and hematopoietic malignancy. SOCS3 is frequently silenced by hypermethylation in gastrointestinal cancers (e.g., in hepatocellular carcinoma[18]and Barrett's adenocarcinoma[19]). Silencing of SOCS3 by promoter methylation in human lung and head and neck cancer has also recently been reported.[20,21]In addition, early studies proposed that DNA methylation and demethylation in the mammalian genome during development are frequent events and play major roles in regulating gene expression and other developmental processes.[22-24]
It is still unknown whether the expression of SOCS3 during the process of liver regeneration is coordinated with changes in DNA methylation; this might be expected because the promoter sequence of the SOCS3 gene is rich in CGIs and its expression in other physiological and physiopathological states is regulated by DNA methylation. In this study, we investigated the alteration of CGI methylation of the SOCS3 gene using a mouse model with PH.
Methods
Animals and treatment
C57BL/6 male mice between 8 and 10 weeks of age were used. Animals were maintained in a specific pathogenfree facility under a 12-hour light/dark cycle with regular chow. The mice were fasted overnight but allowed free access to water prior to treatment. Isoflurane inhalation anesthesia and surgical 2/3 PH were performed as previously described.[25]The animals were sacrificed at the indicated times following surgery to harvest and weigh the remaining liver tissue. A single dose of 5-bromo-2'-deoxyuridine (BrdU) (Sigma-Aldrich, St. Louis, MO) was administered (50 mg/kg i.p.) 30 minutes prior to sacrifice. The body weight and liver weight were recorded. The liver tissues were subjected to DNA purification, paraffinembedded sectioning for pathological examination and BrdU immunohistochemistry. Animal procedures and care were conducted in accordance with the institutional guidelines in compliance with national and international laws and policies.
DNA isolation and detection
Genomic DNA was extracted from the regenerating liver using the Puregene DNA Isolation Kit (Gentra Systems Inc., Minneapolis, MN) according to the product protocol. The purified DNA (5 µL) was subjected to purity assessment and quantification using ultraviolet spectrophotometry.
DNA modification by sodium bisulfate
DNA (2 µg) in a volume of 50 µL was denatured by NaOH (final concentration, 3 mol/L) for 30 minutes at 42 ℃. Thirty microliters of 10 mmol/L hydroquinone (Sigma-Aldrich) and 520 µL of 3.6 mol/L sodium bisulfite (Sigma-Aldrich) at pH 5, both freshly prepared, were added and mixed, and samples were incubated under mineral oil at 50 ℃ for 16 hours. Modified DNA was purified using Wizard DNA purification resin (Promega, Fitchburg, WI) according to the manual and eluted into 50 µL of water. DNA modification was completed by NaOH treatment (3 mol/L) for 15 minutes at room temperature, followed by ethanol precipitation. DNA was resuspended in water and used immediately or stored at -20 ℃.
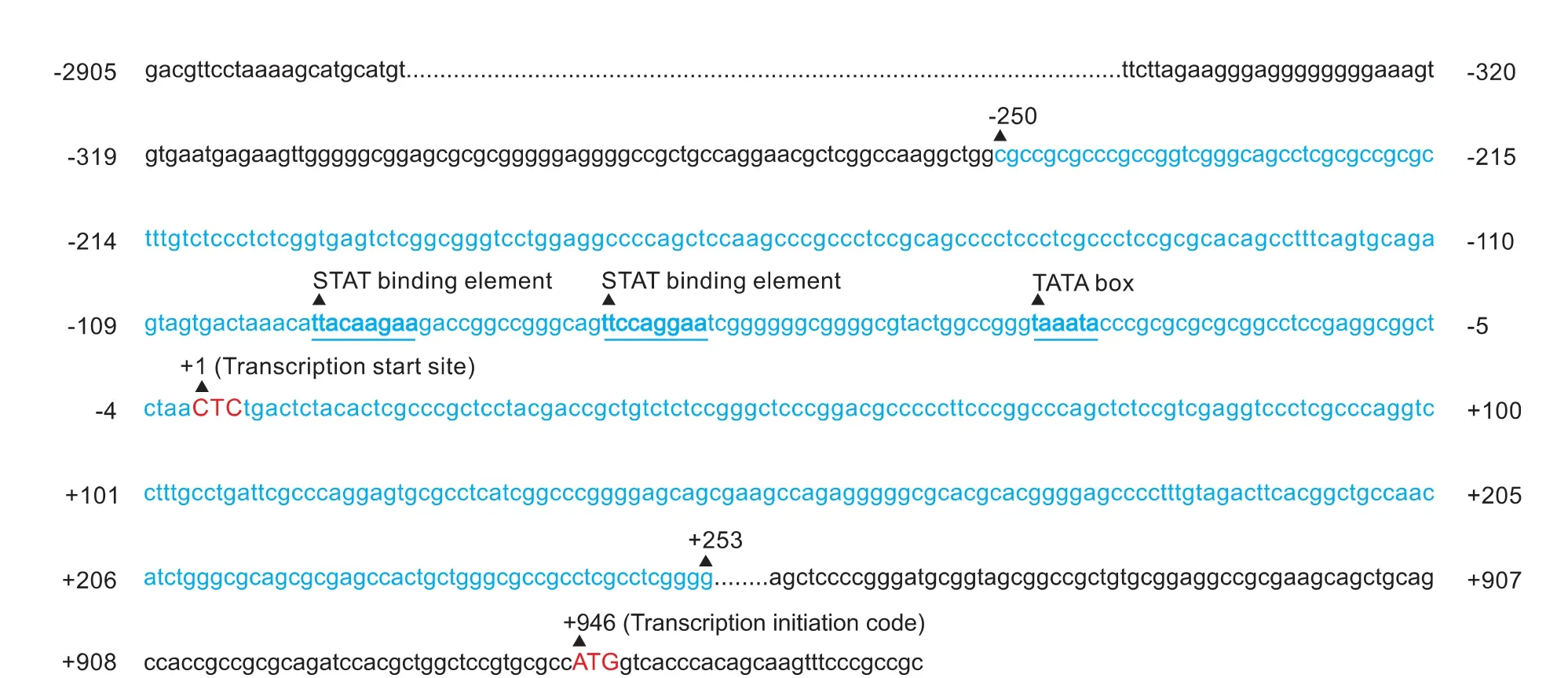
Fig. 1. CGI-rich region of the mouse SOCS3 gene identified by Methyl Primer Express v1.0. CGIs are located in the -250bp to +253bp region.
Primer design and PCR amplification
The complete sequence of the mouse SOCS3 gene in the NCBI database (GenBank sequence number NT 165773.2) was input to Methyl Primer Express v1.0 to identify the CpG-rich region and design the primers.[26,27]The CGI-rich region is located between -250bp and +253bp (relative to the transcription initiation site CTC) in the SOCS3 gene (Fig. 1). In our experiments, primers were designed to amplify the full length of the CGI-rich region and included 58 CGIs. The forward primer was: 5'-GGG GAA AGT GTG AAT GAG AAG T-3', and the reverse primer was: 5'-CCC CTC CTT CCT ACC TAA TCC-3'. The PCR reaction was performed in a final volume of 25 µL containing 1 µL template DNA, 100 nmol/L of each primer, 5 mmol/L dNTP, 2.5 U Taq DNA polymerase (BLR), 2.5 µL 10× PCR buffer and 18 µL sterile deionized water. After denaturing for 5 minutes at 95 ℃, amplification was performed by 30 cycles of 30 seconds at 95 ℃, 30 seconds at 60 ℃, and a combined annealing/extension step of 30 seconds at 72 ℃, using an ABI prism 7000 (PE Applied Biosystems, Darmstadt, Germany). The PCR products were directly loaded onto 1% agarose gel, stained with GoldView, and directly visualized under UV illumination. After PCR, the product was retrieved and inserted into T vector and transfected intoE. colifor positive clone selection and sequencing.
DNA transformation and amplification
The PCR product was ligated into pGEM-T easy Vector (Promega) followed by transformation into competent DH5-αE. colicells. TheE. colicells were plated out onto Lysogeny broth (LB) medium with ampicillin (100 µg/mL). Transformants were identified by the different colors of the colonies and the single white colonies beside the blue ones were selected and amplified in 8 mL LB medium for DNA sequencing.[27]
Western blotting

Fig. 2. Expression of SOCS3 in the process of liver regeneration and reconstitution. A: BrdU incorporation shows DNA replication in the remnant hepatocytes. The positive cells in 1000 counted hepatocytes are presented. B: Ratio of liver weight/body weight after PH. C: Immunoblotting of SOCS3 during the process of liver regeneration; β-actin was used as loading control.
The regenerating liver tissues were harvested at the indicated times from 15 minutes to 7 days after operation for immunoblotting analysis of the protein levels of SOCS3. Total protein was extracted and the protein concentration was determined by the Bradford method (Bio-Rad). Total protein was separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and then transferred to PVDF membrane. The membrane was incubated with 1:200 diluted antibodies against SOCS3 (sc-7009; Santa Cruz Biotechnology, CA) and β-actin was used as loading control. HRP-labelled second antibodies were added and the blotting results were detected by SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Statistical analysis
Data were expressed as mean±standard deviation. Statistically significant differences between the groups were determined by one-way analysis of variance using SPSS13.0 software. APvalue less than 0.05 was considered statistically significant.
Results
Liver regeneration and reconstitution after 2/3 PH
Three of the mice at each indicated time point were sacrificed for liver tissue harvest. The residual liver was removeden bloc, weighed, and the ratio of liver weight to body weight calculated. During the first 24 hours after surgery, the remnant liver weight remained at a low level. In BrdU immunohistochemistry, a few labeled hepatocytes emerged. At 48 hours, the BrdU index was robustly increased and the ratio of liver weight to body weight was nearly double that immediately after operation. Five days after operation the liver was completely reconstituted (Fig. 2A, B).
SOCS3 expression after PH

Fig. 3. CGIs of the SOCS3 gene are irregularly methylated during the process of liver regeneration. A: Schematic of the interpretation of CGI methylation by bisulfite sequencing PCR: after bisulfate treatment, all the cytosines in the non-methylated CpGs are altered to thymines; however, the methylated cytosines remain as cytosines. Through PCR and DNA sequencing, the methylated cytosines were identified when compared to DNA not modified by bisulfate. B: Global view of the methylation frequency of each CGI. C: Variation of the total methylation level at each time point after PH. D: Methylation ratio of each CGI at each time point.
To evaluate SOCS3 protein induction, whole-cell lysates were prepared from liver tissue at the indicated time points after surgery. Increased SOCS3 protein levels were first detected at 30 minutes, with sustained expression for 8 hours. Then, the protein expression decreased precipitously to undetectable levels; however, at 48 hours marked SOCS3 expression reappeared (Fig. 2C). It is thought that some of the newly-regenerated hepatocytes re-enter mitosis immediately after the first cell cycle; this is precisely controlled by molecular pathways, so it is understandable that there might be a second peak of SOCS3 expression. To our knowledge, this is the first report of the changes in SOCS3 expression throughout liver regeneration.
SOCS3 gene not regularly methylated during liver regeneration
To investigate CGI methylation in the SOCS3 gene, we performed bisulfite sequencing PCR and DNA sequencing. In our experiments, the PCR was successfully performed in all specimens. The PCR product was inserted into T vector and transfected intoE. colifor sequencing. Twenty positive clones were randomly selected and amplified in LB medium. After bisulfate treatment, all the cytosines in the nonmethylated CpG are changed to thymines; however, the methylated cytosines remain as cytosines. Through PCR and DNA sequencing, the methylated cytosines were identified when compared to the DNA not modified by bisulfate (Fig. 3A). In total, 30 of 58 CGIs (-247, -241, -234, -201, -160, -139, -132, -130, -83, -55, -41, -29, -25, -21, -14, -9, 28, 32, 51, 114, 126, 134, 149, 164, 168, 196, 214, 219, 244, and 249, relative to the transcription initiation site CTC) were found to be variously methylated before or after operation, and the other 28 remained unmethylated (Fig. 3B). Unexpectedly, only an irregular tendency of the alteration was revealed (Fig. 3C). At each time point, there was a total of 1160 CGIs in the 20 selected clones; however, the number of methylated CGIs ranged between 0 and 16, and the methylation remained at a very low level. In addition, there seemed to be no stable increases and decreases of the methylated CGIs during the process of regeneration. In each of the 30 CGIs with various methylation status, the frequency of methylated sites ranged from 0 to 5 (0-25% of the 20 selected clones) at each time point. The CGI at nucleotide 51 relative to CTC had the highest frequency of methylation, and the ratio was 10% before operation and 15%, 10% and 25% at 1, 2 and 24 hours after PH, respectively. Nucleotide 114 had the lowest frequency of methylation, and only one clone was found to have been methylated at 2 hours after PH (Fig. 3D).
Discussion
In this study the time-course changes were investigated in the expression of SOCS3 throughout liver regeneration after PH, indicating that they were not regulated by changes in the methylation status of CGIs. As a downstream molecule of STAT-3 and participating in a negative-feedback loop to decrease STAT-3 activity, SOCS3 is thought to be the major SOCS familymember induced after PH.[14,15]A study[15]reported that the induction of SOCS3 mRNA and protein levelsis transient, and increasing SOCS3 protein and gene levels are first detected by 2 hours with sustained expression until 8 and 10 hours. Given that SOCS3 plays an essential role in controlling the expression of acute-phase responses, in order to balance the growthpromoting and growth-inhibiting pathways, studies have focused on the transient and immediate activation of SOCS3 protein after PH. However, SOCS3 acts as a suppressor to terminate regeneration; the late effect of SOCS3 in the process of liver regeneration is still unclear, since some hepatocytes re-enter the cell cycle. That is to say, the growth-promoting and growth-inhibitory pathways might be reactivated after the termination of the first cell cycle. In our experiments, SOCS3 was markedly induced immediately after PH but notably depressed at 8 hours. However, another peak emerged 48 hours after surgery. More interestingly, the expression abundance was not significantly less than that at the first episode between 1 and 8 hours. Normally, DNA replication in the residual mouse hepatocytes reaches a peak at 48 hours after PH, which is also consistent with our results. It is rational to speculate that some of the newly-regenerated hepatocytes re-enter the cell cycle, so the reactivation of SOCS3 might also participate in the late cellular and molecular events of liver regeneration.
DNA methylation plays an important role in regulating gene expression, cell proliferation, differentiation, growth and genetic imprinting.[16,17,28,29]Many early studies confirmed that CGIs are the sites of transcriptional initiation, and that they act as promoters in mammalian genomes.[30]Silencing of genes is thought to be either due to direct inhibition of transcriptionfactor-binding by DNA methylation, or by methyl-binding domain proteins that recruit chromatin-modifying activity to methylated DNA.[31]Early studies revealed that changes in DNA methylation in the mammalian genome during development are frequent events and play major roles in regulating gene expression and other developmental processes.[32]CGI methylation has also been well documented in epithelial and non-epithelial malignancies, including hepatocellular carcinoma.[18]The CGIs of several tumor-suppressor genes acquire cancerspecific methylation, and these changes are thought to contribute to uncontrolled proliferation and thus tumor development.[18-21]
Frequent hypermethylation of the functional SOCS3 promoter region has been found to correlate with silencing of the SOCS3 gene in many malignancies including hepatocellular carcinoma and Barrett adenocarcinoma.[18,19]In addition, in low- and high-grade neoplasia as well as in Barrett mucosa, SOCS3 is methylated and shows increasing rates of methylation with higher grades of neoplasia.[19]Accumulating evidence indicates that SOCS3 plays a crucial role both in the regulation of liver regeneration and inhibition of tumorigenesis. However, it is unclear whether CGI methylation participates in the regulation of SOCS3 expression during liver regeneration. In this study, we performed bisulfitesequencing PCR to investigate the detailed nucleotide methylation in the promoter region of the SOCS3 gene. Within the sequence of the promoter region, 58 CGIs were identified and 30 were found to be variously methylated before or after operation. Unexpectedly, all 30 CGIs were irregularly and sparsely methylated. We did not find any time point that presented an extremely high or low ratio of methylated CGIs. In addition, none of the 30 CGIs was regularly methylated throughout the course of regeneration. It seems that all the CGIs are randomly methylated under normal conditions, and the methylation level remains at a very low level. None of our data indicated that the methylation level changed during regeneration. Gene methylation and demethylation act as an important mechanism to regulate transcription, which is critical to development and tumorigenesis, and intervention in DNA methylation often results in abnormal development or the inhibition of tumor proliferation.[33,34]However, intervention in the expression of a particular gene during a routine acute pathophysiological process via DNA methylation regulation often results in a negative outcome.[35]Based on our data, it seems unlikely that liver regeneration is promoted by down-regulating SOCS3 transcription through the induction of DNA regulation.
Contributors:SYJ proposed the study. GG performed the DNA purification and modification. XJ and BJ performed the animal surgery and pathologic analysis. SHQ analyzed the data. All authors contributed to the design and interpretation of the study and to further drafts. BH is the guarantor.
Funding:This work was supported by grants from the Natural Science Foundation of China (30870983 and 30971118).
Ethical approval:Animal procedures and care were conducted in accordance with the institutional guidelines in compliance with national and international laws and policies.
Competing interest:No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
1 Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology 2006;43:S45-53.
2 Michalopoulos GK. Liver regeneration. J Cell Physiol 2007;213: 286-300.
3 Madrahimov N, Dirsch O, Broelsch C, Dahmen U. Marginal hepatectomy in the rat: from anatomy to surgery. Ann Surg2006;244:89-98.
4 Locker J, Tian J, Carver R, Concas D, Cossu C, Ledda-Columbano GM, et al. A common set of immediate-early response genes in liver regeneration and hyperplasia. Hepatology 2003;38:314-325.
5 Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol 2004;5:836-847.
6 Paranjpe S, Bowen WC, Tseng GC, Luo JH, Orr A, Michalopoulos GK. RNA interference against hepatic epidermal growth factor receptor has suppressive effects on liver regeneration in rats. Am J Pathol 2010;176:2669-2681.
7 Yang X, Guo M, Wan YJ. Deregulation of growth factor, circadian clock, and cell cycle signaling in regenerating hepatocyte RXRalpha-deficient mouse livers. Am J Pathol 2010;176:733-743.
8 Cressman DE, Greenbaum LE, DeAngelis RA, Ciliberto G, Furth EE, Poli V, et al. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science 1996; 274:1379-1383.
9 Cressman DE, Diamond RH, Taub R. Rapid activation of the Stat3 transcription complex in liver regeneration. Hepatology 1995;21:1443-1449.
10 Sakuda S, Tamura S, Yamada A, Miyagawa J, Yamamoto K, Kiso S, et al. Activation of signal transducer and activator transcription 3 and expression of suppressor of cytokine signal 1 during liver regeneration in rats. J Hepatol 2002;36: 378-384.
11 Naka T, Narazaki M, Hirata M, Matsumoto T, Minamoto S, Aono A, et al. Structure and function of a new STAT-induced STAT inhibitor. Nature 1997;387:924-929.
12 Endo TA, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K, et al. A new protein containing an SH2 domain that inhibits JAK kinases. Nature 1997;387:921-924.
13 Hilton DJ, Richardson RT, Alexander WS, Viney EM, Willson TA, Sprigg NS, et al. Twenty proteins containing a C-terminal SOCS box form five structural classes. Proc Natl Acad Sci U S A 1998;95:114-119.
14 Yang XP, Schaper F, Teubner A, Lammert F, Heinrich PC, Matern S, et al. Interleukin-6 plays a crucial role in the hepatic expression of SOCS3 during acute inflammatory processes in vivo. J Hepatol 2005;43:704-710.
15 Riehle KJ, Campbell JS, McMahan RS, Johnson MM, Beyer RP, Bammler TK, et al. Regulation of liver regeneration and hepatocarcinogenesis by suppressor of cytokine signaling 3. J Exp Med 2008;205:91-103.
16 Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 2006;31:89-97.
17 Blewitt ME, Vickaryous NK, Paldi A, Koseki H, Whitelaw E. Dynamic reprogramming of DNA methylation at an epigenetically sensitive allele in mice. PLoS Genet 2006;2:e49.
18 Niwa Y, Kanda H, Shikauchi Y, Saiura A, Matsubara K, Kitagawa T, et al. Methylation silencing of SOCS-3 promotes cell growth and migration by enhancing JAK/STAT and FAK signalings in human hepatocellular carcinoma. Oncogene 2005;24:6406-6417.
19 Tischoff I, Hengge UR, Vieth M, Ell C, Stolte M, Weber A, et al. Methylation of SOCS-3 and SOCS-1 in the carcinogenesis of Barrett's adenocarcinoma. Gut 2007;56:1047-1053.
20 Radhakrishnan R, Kabekkodu S, Satyamoorthy K. DNA hypermethylation as an epigenetic mark for oral cancer diagnosis. J Oral Pathol Med 2011 Jun 8.
21 Ho CM, Lai HC, Huang SH, Chien TY, Lin MC, Chang SF. Promoter methylation of sFRP5 in patients with ovarian clear cell adenocarcinoma. Eur J Clin Invest 2010;40:310-318.
22 Senner CE. The role of DNA methylation in mammalian development. Reprod Biomed Online 2011;22:529-535.
23 He XJ, Chen T, Zhu JK. Regulation and function of DNA methylation in plants and animals. Cell Res 2011;21:442-465.
24 Geiman TM, Muegge K. DNA methylation in early development. Mol Reprod Dev 2010;77:105-113.
25 Boyce S, Harrison D. A detailed methodology of partial hepatectomy in the mouse. Lab Anim (NY) 2008;37:529-532.
26 Du P, Ye HR, Gao J, Chen W, Wang ZC, Jiang HH, et al. Methylation of PTCH1a gene in a subset of gastric cancers. World J Gastroenterol 2009;15:3799-3806.
27 Li LC. Designing PCR primer for DNA methylation mapping. Methods Mol Biol 2007;402:371-384.
28 Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis 2000;21:461-467.
29 Delaval K, Feil R. Epigenetic regulation of mammalian genomic imprinting. Curr Opin Genet Dev 2004;14:188-195.
30 Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev 2011;25:1010-1022.
31 Curradi M, Izzo A, Badaracco G, Landsberger N. Molecular mechanisms of gene silencing mediated by DNA methylation. Mol Cell Biol 2002;22:3157-3173.
32 Liang P, Song F, Ghosh S, Morien E, Qin M, Mahmood S, et al. Genome-wide survey reveals dynamic widespread tissuespecific changes in DNA methylation during development. BMC Genomics 2011;12:231.
33 Strathdee G, Brown R. Aberrant DNA methylation in cancer: potential clinical interventions. Expert Rev Mol Med 2002; 4:1-17.
34 Li QX, Yu DH, Liu G, Ke N, McKelvy J, Wong-Staal F. Selective anticancer strategies via intervention of the death pathways relevant to cell transformation. Cell Death Differ 2008;15:1197-1210.
35 Pillich RT, Scarsella G, Risuleo G. Regeneration and DNA demethylation do not trigger PDX-1 expression in rat hepatocytes. World J Biol Chem 2010;1:281-285.
July 28, 2011
Accepted after revision December 20, 2011
Author Affiliations: Key Laboratory of Transplant Engineering and Immunology, Ministry of Health (Guo G, Xia J, Bao J, Sun HQ, Shi YJ and Bu H) and Department of Pathology (Bu H), West China Hospital, Sichuan University, Chengdu 610041, China
Yu-Jun Shi, MD, PhD, Key Laboratory of Transplant Engineering and Immunology, Ministry of Health, West China Hospital, Sichuan University, Chengdu 610041, China (Tel: 86-28-85164030; Fax: 86-28-85164034; Email: shiyujun@scu.edu.cn)
© 2012, Hepatobiliary Pancreat Dis Int. All rights reserved.
10.1016/S1499-3872(12)60198-2
杂志排行
Hepatobiliary & Pancreatic Diseases International的其它文章
- Gastric- and intestinal-type marker expression in invasive ductal adenocarcinoma of the pancreas
- Early changes of hepatic hemodynamics measured by functional CT perfusion in a rabbit model of liver tumor
- A common variant in the precursor miR-146a sequence does not predispose to cholangiocarcinoma in a large European cohort
- Muscarinic acetylcholine receptor M3 in proliferation and perineural invasion of cholangiocarcinoma cells
- Hepatocyte differentiation of mesenchymal stem cells
- Early control of short hepatic portal veins in isolated or combined hepatic caudate lobectomy
