Study on Classification and Genetic Diversity of Kentucky Bluegrasses by Using RAPD Markers
2012-07-02WangChaoLiXinGaoLinaZhangLuLiuWeiLiuHuiminandChenYajun
Wang Chao, Li Xin, Gao Li-na, Zhang Lu, Liu Wei, Liu Hui-min, and Chen Ya-jun
College of Horticulture, Northeast Agricultural University, Harbin, 150030, China
Introduction
Kentucky bluegrass (Poa pratensis L.)is one of the most widely used cool-season turf grasses in the world. This species originated in Europe, north of Asia and Africa, before it was introduced in North America. The USA had improved on the Kentucky bluegrass since 1950s, and gradually industrialized varieties worldwide. China introduced many Kentucky bluegrass varieties from abroad in recent 20 years,which enriched the turfgrass cultivar resources in China, but at the same time, resulted in the degradation of those that couldn't adapt to the local climate environment. Because of the blind introduction of some cultivars and unfamiliarity with their genetic background and bionomics, serious economic loss ensued. In view of this, it's now necessary to make taxonomic research on the introduced Kentucky bluegrass cultivars and to understand the genetic relationship and the genetic background among different cultivars.
This study adopted the introduced European and American Kentucky bluegrass varieties in addition to native materials from China, and then the genetic diversity was analyzed by using the RAPD (Random Amplif i ed Polymorphic DNA)analysis. The objective of this study was to classify Kentucky bluegrass cultivars to estimate the genetic diversity and variety of range in cultivars, and also to detect the genetic relationship among different Kentucky bluegrass cultivars by RAPD markers, which would provide theatrical basis for further reasonable utilization of introduced Kentucky bluegrass cultivars and the development of domestic Kentucky bluegrass resources and breeding work in China.
Materials and Methods
Materials
The materials of Kentucky bluegrass were introduced from Europe and America, and one wild cultivar origined from Heilongjiang, China. The cultivars code,name and origin area are shown in Table 1.
Methods
DNA extraction methods
Every sample was randomly selected from three plants and their tender leaves. We abstracted con-genome DNA by using SDS methods and then detected the integrality and density of DNA.
The RAPD methods
The reaction system of the RAPD was as the followings: 1 μL template DNA (10-20 ng · μL-1), 2 μL 10× reaction buffer, 1.8 μL MgCl2(25 mmol · L-1), 0.4 μL dNTP (2.5 mmol · L-1), 0.2 μL TaqDNA polymerase(5 U · μL-1), 1.5 μL RAPD primer (10 pmol · μL-1),added deionized water until reaching 20 μL. The RAPD primer was synthesized by Bioasia.
The reaction process of the RAPD was as the followings: 94℃ fore-degeneration 5 min, 94℃ degeneration 30 s, 37℃ annealing 1 min, 72℃ extention 1 min,30 s circulation 34, 72℃ extention 7 min, then stopped at 4℃. After staining with bromphenol blue, PCR production was electrophoresised for 1.5 h in 1.5%(w/v)sepharose. Pictures were taken using the gelatin system after electrophoresis, and then the data was recorded. Some of the bad results or those with missing primers and samples should be electrophoresis and amplif i ed.

Table 1 Kentucky bluegrass cultivars and their origins
Data processing
In electrophoresis map, we represented a site where primer and template DNA were complementary, which a molecular marker estimated the molecular weight of amplified product according to the corresponding location of molecular weight in the gelatin and on the basis that the weight of DNA was inversely proportional with electrophoresis mobility on the range of resolution.
Electrophoresis of the same primer and different templates showed that the RAPD band with the same mobility rate had the same molecular weight. Bands presented in all the templates were public bands,expressing no polymorphism, while others unique to the template were polymorphic bands, expressing polymorphism. The RAPD was dominant marker,the individuals with the bands were marked 1, those without were marked 0, and those that had lost the bands were marked -1.
Genetic parameters calculation:
The similar coefficients between germplasm were simple matching coefficient (SM), SM=(a+d)/(a+b+c+d). a was the amount when both of the compared varieties were 1, which was called correct matching; b and c respectively indicated the amount when compared varieties were 1 and 0, which was called incorrect matching; d was the amount when compared varieties were -1, which was called negatively matching.
The similar coeff i cients used in the experiment were as the followings:
Jacquard's coeff i cient (JACCA, 1908):
Jcij=a/(a+b+c)
Jcij represented the genetic similar coefficients between i and j in this formula, a was the quantitative band within two materials, b was the band's amount that was within material i but not in material j, c was the band's amount that was within material j but not in i.
Results
Primer screening
Took four different Kentucky bluegrass varieties as templates, used the best reaction system to carry out PCR amplif i cation to 500 item primers, and screened primers according to the amplification strap results.The principle was that amplification strap of primer was more, clear and having polymorphism.
RAPD polymorphism analysis
This experiment proceeded to polymorphism screening of 500 random primers made from Bioasia Company, 431 primers (account 86.2%)had not produced products which could be detected. A total of 69 primers could produce polymorphism band, and we could only choose 46 primers to amplify to get clear amplif i cation bands. The results are shown in Figs. 1 and 2.

Fig. 1 The RAPD map amplif i ed by U03 primer
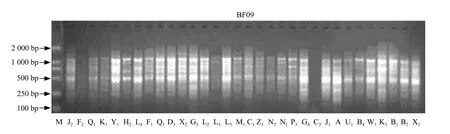
Fig. 2 The RAPD map amplif i ed by BF09 primer
From Figs. 1 and 2 we knew that every primer got genome DNA finger printing, the polymorphism behaved differently with a different primer, and different primers showed significant differences with different materials' finger printings and amounts of amplif i ed bands; the same primers had different fi nger printing with different materials. Amplified straps were tested from different primers ranging from 1 to 9, with every primer amplifying 4.3 strap on average.The primers which amplified the most straps were BC10, W12, X02 etc.; the primers which amplified the fewest straps were U14, BB17, V15, and W20.Forty-six primers totally produced 197 different molecular weight RAPD markers in 32 samples,among which mono-morphs bands were two, which accounted for 1.01%, polymorphism bands were 195,which accounted for 98.5%. It explained the genetic background complexity among Kentucky bluegrass cultivars.
Genetic relationships of cultivars
Pair-wise similarity coefficient of Jaccard cultivars'coefficient of similarity was between 0.125 and 0.714, the result showed that there was significant heterogeneity difference among different cultivars; the highest pairs of cultivars similar coefficient were O1and X2; the highest average coefficient of similarity was H2, the lowest one was C2, it indicated that H2had the minimum genetic distance to the other cultivars,and had the closest genetic relationships; C2had the maximum genetic distance to other cultivars, therefore,the farthest genetic relationship.
Utilizing NTSYS-PC2.11a software, data was clustered according to 1, 0, -1 which inverted from the RAPD amplif i cation results. Jaccard amplif i cation results are shown in Fig. 3 and Table 2.

Fig. 3 Statistics results of Jaccard genetic similarity among 32 Kentucky bluegrass cultivars by the RAPD
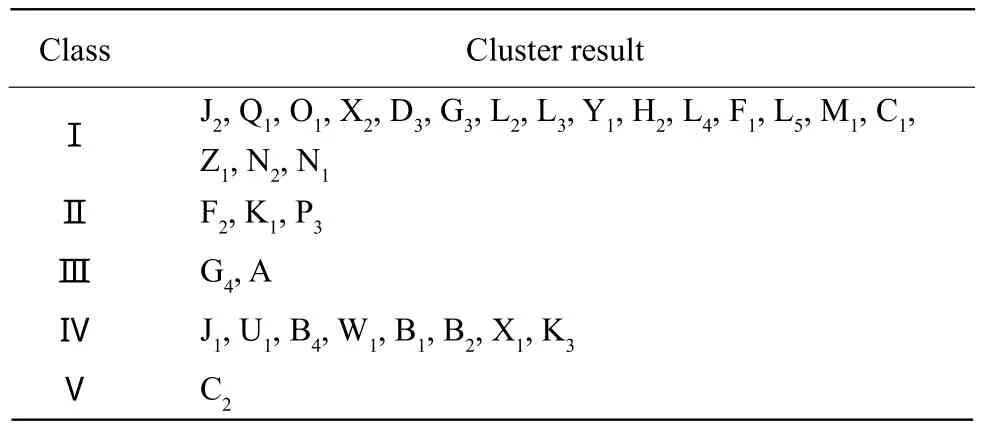
Table 2 Cluster result of 32 Kentucky bluegrass cultivars by the RAPD
We could see from Fig. 3 that the experimental samples were divided into fi ve groups with coeff i cient similarity of 0.434: the first group included 18 cultivars: J2, Q1, O1, X2, D3, G3, L2, L3, Y1, H2, L4, F1,L5, M1, C1, Z1, N2, and N1; the second group included three cultivars: F2, K1, and P3; the third group included two cultivars: G4and A; the fourth group included eight cultivars: J1, U1, B4, W1, B1, B2, X1, and K3; K3and B1, two cultivars from Denmark, illustrated a closer relationship to these six American cultivars; the fifth group included one cultivar: C2. In 32 Kentucky bluegrass cultivars, there were 18 cultivars in group 1, eight cultivars in group 4, the remaining six cultivars were in the other three groups, which illustrated that the genetic relationship among European and American introduced varieties was relatively closer and hereditary basis was much narrower.The coefficient similarity between A (Heilongjiang Province native wild cultivars)and other cultivars was not the smallest, showing that A had some genetic relationships with European and American cultivated varieties, which meant there were Chinese genes in European and American cultivated varieties.
Conclusions
This study set up the best RAPD reaction system and reaction condition which suited Kentucky bluegrass.Genetic similarity computation results indicated that there was significant heterogeneity among different cultivars: 32 cultivars were distributed to fi ve groups.The genetic relationship of European and American introduced varieties was closer and hereditary basis was much narrower. The morphological marker method was used as the main method of identifying the authenticity and purity of cultivars. With the increasing in the production and development of Kentucky bluegrass cultivars, the morphological difference among different cultivars was becoming smaller and smaller, making it very difficult to distinguish them solely from morphology. Using the RAPD marker along with other markers to study genetic diversity of germplasm resources of Kentucky bluegrass to identify varieties would be more accurate, fast, simple and plentiful; therefore, the RAPD is an effective method for variety classif i cation and identif i cation.
Chen Z T, Huang Y B, Ying Z Y, et al. 2008. Application of RAPD to Pennisetum., a forage resource. Chinese Journal of Tropical Agriculture, 28(4): 7-10.
Greens S L, Hart T C, Afonm A. 1999. Using geographic information to acquire wild crop germplasm for excollections (II): post-collection analysis. Crop Sci, 39: 843-849.
Johnson R C, Johnston W J, Golob C T. 2002. Characterization of the USDA Poa pratensis collection using RAPD markers and agronomic descriptors. Genetic Resources and Crop Evolution, 49: 349-361.
Li X L, Yu Z, He P C, et al. 2008. RAPD analysis of Elymus canadensis, E. excelsus and their hybrid F_1. Acta Agrestia Sinica,16(1): 23-27.
Li Y C. 2002. RAPD analysis on inter-species relationships in Poa. Acta Pratacultural Science, 11(4): 94-99.
Stammers M. 1995. Use of random PCR (RAPD)technology to analyse phylogenetic relationships in the Lolium/Festuca complex. Heredity,74(1): 19-27.
Tian Y, Wang C. 2008. Initial analysis on genetic relationship of Brassica oleracea L. by RAPD. China Vegetables, 1: 20-22.
Zhang H Y, Wang Y J, Xu Y, et al. 1998. RAPD used in analysis of genetic relationships of cucumber (Cucumis sativus L.)germplasm.Acta Horticulturae Sinica, 25(4): 345-349.
Zhong H Q, Wu J S, Chen S L, et al. 2009. Genetic diversity analysis of ornamental sunf l ower germplasm resources with RAPD. Molecular Plant Breeding, 7(1): 73-78.
杂志排行
Journal of Northeast Agricultural University(English Edition)的其它文章
- Establishment of TaqMan Real-time Quantitative PCR Assay for Foreign Gene Copy Numbers in Transgenic Soybean
- Study on Semen Freezing Preservation of German Shepherd Dogs
- In Vitro Study on Commercial Organic Acid Activate WD (WD)Against Four Pathogenic Bacteria
- Response of Osmotic Adjustment of Lactobacillus bulgaricus to NaCl Stress
- Temperature Simulation of Berry Slices Under Microwave Vacuum Puff i ng Conditions
- Storage System Design Scheme in Virtualization Construction
