DNA Methylation Profiles of Protease Nexin 1 (SERPINE2) Gene in Human Cell Lines
2011-07-18ShanGaoPeterAndreasen
Shan Gao, Peter A. Andreasen
Danish-Chinese Centre for Proteases and Cancer, Department of Molecular Biology, University of Aarhus, 8000 Aarhus C, Denmark
DNA Methylation Profiles of Protease Nexin 1 (SERPINE2) Gene in Human Cell Lines
Shan Gao*, Peter A. Andreasen
Danish-Chinese Centre for Proteases and Cancer, Department of Molecular Biology, University of Aarhus, 8000 Aarhus C, Denmark
Objective:To investigated whether epigenetic mechanisms contribute to the variable expression of variable protease nexin1(PN-1) encoded by theSERPINE2gene in different cell types.
Methods:Working with 5 human cell lines, we determined the CpG methylation status within two CpG islands in theSERPINE2gene by bisulphate sequencing and the PN-1 mRNA level by Q-RT PCR.
Results:A CpG island spanning the transcription initiation site showed little methylation in 3 of the cell lines and substantial methylation in 2 of the cell lines. A CpG island covering the translation starting site showed full methylation in all investigated cell lines. Methylation within the CpG island was not randomly distributed, but showed accumulation at specific sites. However, we were not able to distinguish any patterns which related the methylation frequency to the gene expression level. Inhibition of CpG methylation with 5-aza-2’-deoxycytidine led to a several fold increase in PN-1 mRNA levels, but based on the results on CpG methylation in the CpG island spanning the transcript, the effect is most likely indirect.
Conclusion:We have carefully mapped the CpG methylation pattern in two CpG islands in the 5’ part of theSERPINE2gene without finding any obvious inverse correlation between methylation frequency and expression level.
Protease nexin 1;SERPINE2; DNA methylation; Cancer
INTROCUTION
There is an increasing interest inhibitors as prognostic or predictive markers in human cancers. This is particularly true in the case of plasminogen activator inhibitor-1 (PAI-1). PAI-1 is a fast and specific inhibitor of the urokinase-type plasminogen activator (uPA), a serine protease which catalyses the conversion of the inactive zymogen plasminogen to the broad-spectrum serine protease plasmin. Both uPA and PAI-1 are strongly implicated in cancer invasion and metastasis, as a finely tuned uPA-catalysed plasmin generation in tumours is believed to facilitate cancer cell migration through extracellular matrix and basement membranes. For instance, high tumour levels of uPA and PAI-1 in tumour extracts are markers for a poor prognosis in several human cancer types[1-4]. In fact, the tumour levels of uPA and PAI-1 are now recommended by the American Society for Clinical Oncology as tumour markers in the management of breast cancer[5]. Also, the level of type-1 111111tissue inhibitor of metalloproteases (TIMP-1) in tumour extracts and blood plasma is suggested as a prognostic marker in lung, kidney, and breast cancers as well as lymphoblastic leukemia[6-11].
A less studied protease inhibitor, protease nexin 1 (PN-1), encoded by theSERPINE2gene, is a fast inhibitor of several serine proteases, including thrombin, plasmin, urokinase-type plasminogen activator, and matriptase. PN-1 is the closest phylogenetic relative of PAI-1 and also belongs to the serpin superfamily. PN-1 has important neurobiological functions, but recent studies have indicated important tumour biological functions of PN-1. Overexpression of PN-1 was found in the majority of the investigated pancreatic, gastric and colorectal cancers[12,13], breast carcinomas[14]as well as oral carcinomas[15]. In oral carcinoma, it was shown that an increased PN-1 level was correlated with positive lymph node metastasis[15]. On the basis of studies with a mouse model, PN-1 was also suggested as a candidate promoter for lymph node metastasis of testicular cancer[16].
In tumours, protease inhibitors may be expressed by a variety of cell types. Thus, in some cases, PAI-1 is expressed by stromal fibroblasts[17-19], but in oral squamous cell carcinomas, it is expressed by the malignant cells themselves[20,21]. However, the PN-1 expressing cell types inhuman cancers have not been identified yet. Only one study indicates that PN-1 is exclusively expressed by tumour cells in pancreatic cancers, whereas the duct cells, the acinar cells and the stromal cells of the adjacent normal area are completely negative for PN-1 expression[12].
Epigenetic mechanisms have been demonstrated in regulation of the gene expression of several serpin genes[22,23].In one study[23], 18 serpin genes were analysed for their methylation profiles in preeclampsia. However, the study did not investigate some other serpin genes, including the PAI-1 (SERPINE1) gene and the PN-1 (SERPINE2) gene, reportedly due to primer or sequencing problems. Our group has previously found that variable methylation of the PAI-1 gene 5’-flanking region is inversely correlated with variable transcription of the PAI-1 gene in different cell lines, and indicated that silencing by methylation may act in conjunction with histone deacetylation[24]. ForSERPINE2, it was previously shown that the promoter activity of the gene in rat could be inhibited byin vitromethylation of the sequence containing AP2 and Sp1 binding sites, within a CpG island around the initial transcription site (spanning from -300 bp to exon I)[25]. But overall, less information about epigenetic mechanisms is available for the humanSERPINE2gene. An elucidation of the epigenetic mechanisms in determining the expression of PN-1 in different cell types is therefore a particularly interesting challenge. In the present study, we designed primers containing CG sites, but using pyrimidines or purines instead of cytosines or guanines. Therefore, it could be used to amplify either methylated or unmethylated alleles and be able to solve the primer problem caused by high density of CpG sites. We investigated the CpG methylation status in five human cells lines derived from different cell types. Our studies suggest that DNA methylation at specific sites may to some extent downregulate PN-1 expression, but that other, as yet unidentified factors are also involved in the regulation of the highly variable expression between different cell lines.
MATERIALS AND METHODS
Cell Culture
The culture of the three human cell lines HT-1080, MCF-7 and U937, including treatment by 5-aza-2’-deoxycytidine (5-Aza-CdR) (Sigma) and trichostatin A (TSA) (Sigma), was described previously[24]. Two other human cell lines, one derived from breast carcinoma, T47D, and one from a T-cell leukemia, Jurkat, were added for the present study and cultured following the same procedures[24].
After harvest, the cells were separated in two parts, one for preparation of DNA (DNeasy Tissue Kit, Qiagen) and one for preparation of cytoplasmic RNA (RNeasy Mini Kit, Qiagen). On-column DNase digestion by RNA-free DNase I (Qiagen) was performed during the RNA purification procedure.
In Silico Analysis for CpG Islands
The genomicSERPINE2sequence (GeneBank Accession No. NM_006216) was screened for CpG islands by the use of two CpG plot programmes (http://www.ebi.ac.uk/ emboss/cpgplot/) and (http://www.ucsf.edu/urogene/ methprimer/index1.html), with the following parameters: 100 bp length, 50% G+C and >0.6 observed/expected ratio. The ratio of observed to expected CpG is calculated according to Gardiner-Garden and Frommer’s formula, Obs/Exp CpG=Number of CpG* N/(Number of C* Number of G), where “N” is the total numbers of nucleotides in the sequence analysed[26].
Bisulphite Sequencing
Bisulphite sequencing was performed as described elsewhere[24]. For a CpG island between bp -539 to bp +776 of theSERPINE2gene, two amplifications were performed. One, covering the 5’ end of the sequence, was amplified by a nested PCR. The outer primers were: PN PRO-F, GTTTTTTTTAGGGGGTAGTTAAG (start at bp -391); and PN PRO-R, AAAATCACAACCRA AAAAAAAC (R as A or G) (end at bp +71). The inner primers were: PN-Fin, AAGAY GATATGTTTTGYG TTTTAGGG (Y as C or T) (start at bp -371); PN-Rin: AAAATCACAACCRA AAAAAAA CAACRA TAAC (R as A or G) (end at bp +71). The annealing temperature was 55°C and 57°C for the primary and the secondary reaction, respectively. For amplification of the 3’part of the same CpG island, a single reaction was performed. The primers were: PN-ExInF, GTTTTTTTTYGGTTG TGATTTT(Y AS C OR T) (start at bp +51); and PN-ExIn R, TACTCCCAACTACRAATATCACTA AAC (R as A or G) (end at bp +507). The annealing temperature was 57°C. The two amplicons cover total 119 CpGs. For amplification of the CpG island in exon 2, the primers were: PN-Ex2-F, TGG AAGGAATTATGAATTGGTATTT (start at bp +37377, the translation starting site ATG is underlined at bp +37388); and PN-Ex2-R, CCTCCTATCACAAAACTCTC AAAC (end at bp +37706). The annealing temperature was 57°C. The amplicon covers 14 CpGs.
Amplified DNA was subcloned into the vector pCR2.1 by using a One Shot Electroporation TOPO TA cloning kit (Invitrogen) and sequenced by AGOWA (Berlin, Germany). At least eight positive clones were analysed for each sample. The frequency of total methylated CpGs (FTM) was calculated as (number of methylated CpGs/total number of detected CpGs) x 100%[24].
Quantitative Real-time RT PCR
Reverse transcription and quantitative real-time RT PCR was performed as described previously[15]. All PN-1 mRNA levels were normalized by the level of GAPDH mRNA and expressed relative to the mRNA level in HT-1080 cell, which has a high PN-1 expression and was run as a positive control in each reaction.
Statistical Analysis
All analyses of effects of 5-Aza-CdR and TSA on PN-1 mRNA by quantitative real-time RT PCR were performed at least three times and presented as ¯x±s. Paired-t test was used for comparing the effects of 5-Aza-CdR and TSA.
RESULTS
Variation of PN-1 mRNA Expression Level between Human Cancer Cell Lines
Quantitative real-time RT PCR was used to determine the PN-1 mRNA level in 5 cell lines. A wide variation (~2000 fold) was found. The highest level was seen in HT1080 cells, while relatively low levels were found in U973, T47D and Jurkat cells. The lowest level was seen in MCF-7 cells. In fact, the PN-1 mRNA level in HT-1080 was about 100 fold higher than in any of the other cells lines (Figure 1).

Figure 1.PN-1 mRNA expression of five cell lines. The PN-1 mRNA levels in the indicated cell lines were determined by real time RT-PCR. In each cell line, the level was normalised against the level of GAPDH mRNA level. The level in HT-1080 cells was arbitrarily set to 100, and the levels in the other cell lines expressed relative to that. The bars showx±sfor 3 determinations.
Localisation of CpG Islands in the SERPINE2 Gene
The humanSERPINE2gene is localised on chromosome 2q36.1. The genomicSERPINE2sequence (GeneBank Accession No. NM_006216) was screened for CpG islands. By screening an interval of 30 kb upstream and the whole sequence downstream from the transcription initiation site, we found one CpG island (CpG island 1) from bp -539 to bp +776 relative to the transcription initiation site, containing 155 CpGs. Three more CpG islands were located in intron 1 (bp 17248 - bp 17395, length 148 bp, 8 CpGs), exon 2 (bp 37474 to bp 37669, length 196 bp, 14 CpGs) and exon 4 (bp 47331 - bp 47469, length 139 bp, 10 CpGs), respectively. The exon 2 CpG island is located downstream of the translation start site ATG at bp 37388 (Figure 2).
Methylation Profiles by Bisulphite Sequencing
In the present study, we analysed the CpG islands located around the transcription initiation site and the translation starting site ATG, which contain more CpG sites than the other CpG islands. Such CpG islands are usually considered to have a potential role in regulation of expression. To investigate if epigenetic mechanisms are involved in regulation of PN-1 expression, bisulphite sequencing was performed to determine CpG methylation in the two CpG islands in the different cell lines.
As the size of the CpG island spanning the transcription initiation site is over 1.3 kb, we used two amplifications for the analyses. The 5’ end of sequence, around the promoter region, was amplified by a nested PCR reaction. The 3’ end spanned from exon 1 into intron 1, the forward primer partly overlapping with the reverse primer of last amplification (Figure 2). Therefore, ~77% (119 of 155) CpGs within this CpG island were analysed in the five cell lines. The methylation status from all individual clones is shown in Figure 3A. Total methylation frequency (FTM) within this region were 15.8% in U937 cells, 5.9% in Jurkat cells, and 2.1% in MCF 7 cells, and less than 1% in T47D and HT1080 cells. Most methylated CpGs in Jurkat was localised in the 3’end of the CpG island (exon1-intron1), and multiple CpG sites were found to be methylated in 4 of 8 clones. In contrast, most methylation in U937 cells was localised in the 5’ end of the CpG island (Promoter) and 4 out of 12 DNA clones showed methylation at almost all CpG sites in the promoter region. Most of methylated CpG sites in MCF-7 were also seen within this region (Figure 3A, B and C). Nevertheless, we did not observe any reverse correlation between expression level and methylation frequency within this CgG island, neither when considering total methylation frequency, methylation in the 5’ end nor methylation in the 3’ end.
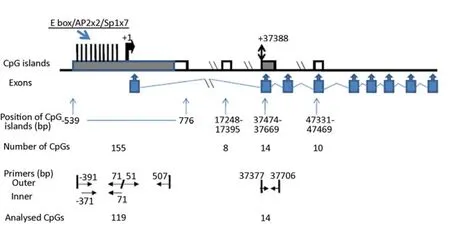
Figure 2.CpG islands in theSERPINE2gene. The transcription initiation site (+1) and the translation starting site (+37388) are indicated. The known transcription factor binding sites at the 5’ end are indicated by thick vertical black bars pointing upwards – 1 E box, 2 AP2 and 7 Sp1 binding sites. Nine exons are indicated by up arrow callouts. Four CpG islands are indicated by boxes; filled box (or filled part of the box) are analysed in the present study and open boxes (or open part of the box) are not. Location of CpG islands, numbers of CpGs within islands, location of primers for bisulphite sequencing, and numbers of analysed CpGs are indicated.
Looking for less obvious reverse correlations between expression and methylation frequency, we focused on several specific CpG sites within this CpG island. In the 5’end of the CpG island, there are 7 Sp1, 2 AP2 binding sites and one E box (Figure 2)[27]. Based on observation with methylation of such sites in the rat SERPINE2 gene[25], methylation of these sites could be hypothesised to be involved in regulation of expression. There are a total of 13 CpGs within or between these sites. In MCF 7 cells, the most frequently methylated CpGs were located at those CpG sites although a low methylation frequency was observed in general. We therefore calculated the methylation frequency at these specific sites. Nine of 13 CpGs are within those Sp1 and AP2 sites(#40,33,32,26,25,24,22,21,20), and 4 between or nearby those sites (#43,42,41,34). The total methylation frequency of the 13 CpGs was 26.3 %, 9.3% and 2.9% in U937 cells, MCF-7 cells and Jurkat cells, respectively, but almost no methylation in HT1080 cells and T47D cells (Figure 3B). Again, we could not find any significantly reverse correlation between expression and methylation frequency.
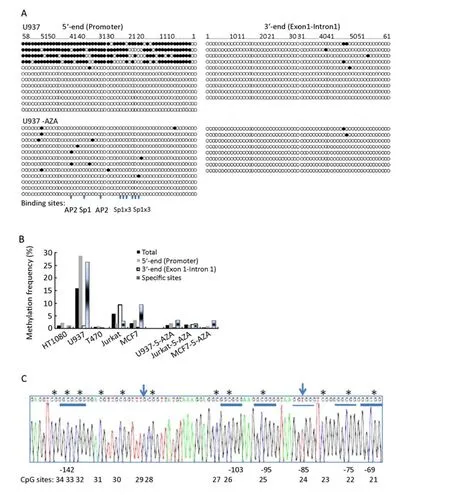
Figure 3.CpG methylation in the CpG island spanning the transcriptional initial site in theSERPINE2gene (bp -539 to bp +776) in human cell lines. A. The methylation status in individual clones among five cells was analysed by bisulphite sequencing. The circles represent the single CpG sites, which are numbered # 58 to 1 from the 5’ end for the amplification of the promoter region and numbered #1 to 61 for the amplification of the exon1-intron1 region. Black circle, methylated; empty circle, unmethylated. The figures show the methylation status for 8-12 single DNA clones for each pair of primers, one for each row. For U937, Jurkat and MCF-7, methylation status of cells with 5-Aza-CdR treatment was included, too. Two AP2 and 7 Sp1 binding sites at the promoter region are also indicated. To control the efficiency of bisulphite treatment, the conversion rate of cytosine at non-CpG sites was calculated based on the alignment of 50 sequences from individual clones. Ten unconverted cytosines was found among total 5400 non-CpG cytosines, therefore the conversion ratio was represented as 99.8%. B. The CpG methylation in the indicated cell lines was analysed by bisulphite sequencing. The frequency of total methylated CpGs (FTM) among 119 of 155 CpG sites was indicated as “Total”; methylation frequency at the “5’ end” and “3' end” was presented according to two amplifications; methylation frequency at 13 specific sites was represented as “specific sites”, as described in the text. The indicated cell lines were cultured under control conditions or for 3 days with 5.0 μmol/L (MCF 7 cells) or 1.0 μmol/L(for U937 and Jurkat cells) 5-Aza-CdR. C. An example shows methylated CpGs within an AP2 (GCGCGGG at bp -142) and 4 Sp1 sites (GGCGGG or GGGCGG at bp -103, -95, -75, -69) [27] at the 5’ end of the Cp*G island spanning the transcription start site (underlined) and CpG sites are also numbered accordingly. Methylated cytosines is marked with “”; two unmethylated CpGs (#29 and #24) is indicated by arrows and CpG #24 is within in one Sp1 site. The location of AP2 and Sp1 was indicated under the sequence.
We also screened the CpG island within exon 2 spanning the translation start site. Almost full methylation for all clones was found in all five cell lines investigated (Figure 4).
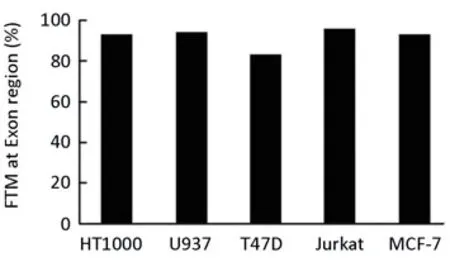
Figure 4.CpG methylation in CpG island spanning the translation start site of the SERPINE2 gene (bp 37474 to bp 37669) in human cell lines. The methylation of 14 CpGs was analysed by bisulphite sequencing. The frequency of total methylated CpGs (FTM) was calculated as described in Materials and Methods.

Figure 5.Effects of 5-Aza-CdR and TSA on the PN-1 mRNA levels in human cell lines. The indicated cell lines were cultured for 3 days with or without 5.0 μmol/L (MCF 7 cells) or 1.0 μmol/L (for U937 and Jurkat cells) 5-Aza-CdR, by which time the culture was continued for an additional day with or without 0.5 μmol/L TSA. After harvest, the PN-1 mRNA levels were determined by real time RT PCR and normalised against the GAPDH mRNA levels. The figures showfor three independent experiments. The results were evaluated by Studentt-test:*significantly different from untreated control,P<0.05;**significantly different from untreated control,P<0.01.
Effect of 5-Aza-CdR and TSA Treatment on PN-1 mRNA Expression
In order to obtain more information about the possible epigenetic regulation of PN-1 mRNA expression level, inhibition of CpG methylation by 5-Aza-CdR and inhibition of histone deacetylase by TSA was performed. The effect of the 5-Aza-CdR treatment on CpG methylation was controlled by bisulphite sequencing, which showed a significantly reduced methylation frequency (Figure 3). An increased PN-1 expression was observed in MCF-7 and U937 cells by both 5-Aza-CdR and TSA treatment, but in Jurkat cells the PN-1 expression level was not increased by any single treatment. An enhancement was observed by combination of both treatments (Figure 5). These results indicated that the CpG methylation and histone acetylation status is implicated in the control of transcription of theSERPINE2gene, but in view of the findings reported above, the effect of 5-Aza-CdR is unlikely to be directly on methylation of CpGs within the CpG island spanning the transcription initiation site.
DISCUSSION
We here report to our knowledge the first study of the CpG methylation patterns in two CpG islands of the human PN-1 gene. The analysis of DNA methylation of the CpG island spanning the promoter region of PN-1 gene has proved difficult because of the high density of CpG sites and the length of the CpG island, i.e., 155 CpGs within 1316 bp length. It is impossible to design primers for bisulfite sequencing following the routine rules, for example, avoiding the CpGs within the primer sequence. We solved these problems by designing primers with pyrimidines (either C or T) for the forward direction and purines (either A or G) for the reverse direction, which allowed amplification of both methylated and unmethylated cytosine[28]. Nested PCR was also applied to improve the amplification efficiency.
Although we are not yet able to draw any clear conclusion concerning the possible involvement of DNA methylation in the CpG island in transcriptional regulation of PN-1 gene, our results do present a first step and provide a basis for further functional studies. It is clear that CpG methylation varies considerably between different cell lines, quantitatively as well as qualitatively. On this basis, we suggest that epigenetic mechanisms may be well involved in transcriptional regulation of the PN-1 gene. Firstly, we found cell line-specific variations in methylation of CpGs localised within the binding sites for several transcription factors which have previously been implicated in regulation of the PN-1 gene[27]. Methylation of the binding sites for these transcription factors has been previously been demonstrated to prevent binding of these factors and to cause downregulation of various genes[29-33], especially in the genes with the first untranslated exon containing a GC rich sequence[34].Also, inhibition of CpG methylation by 5-Aza-CdR caused an increase in PN-1 mRNA level in U937 and MCF-7 cells following a great decrease of methylation frequency also at those specific CpG sites. But since 5-Aza-CdR treatment causes a global inhibition of CpG methylation, we have no clues as to whether methylation of other CpG sites rather than this CpG island may be involved in the regulation of the PN-1 gene and the present data do not give us any reason to believe that the effect of 5-Aza-CdR is directly on the SERPINE2 promoter region. Rather, 5-Aza-CdR may lead to an increased expression of transcription factors involved in regulation of SERPINE2 gene transcription. Also, there was no reason to believe that the CpG island around the start site of translation is involved on transcriptional regulation of the PN-1 gene, since it was almost complete methylated in all cell lines studied. It is clear that factors different from CpG methylation are also important for transcriptional regulation. The variation in CpG methylation and expression level may well be related to the fact that each cell line was derived from different cell types, thus reflecting cellular differentiation. Lack ofmethylation may be a prerequisite for a high transcription rate, but it is certainly not sufficient. For instance, methylation was virtually absent in HT-1080 cells as well as in T47D cells, while these two cell lines still have a more than 100-fold different level of PN-1 mRNA.
The methylation pattern of individual clones shows that methylation may vary strongly among individual cells in the same cell line. In the future, it will therefore be important to be able to correlate expression level and methylation on a single cell basis.
Human SERPIN (serine protease inhibitors) genes encode over 36 proteins with complex conserved structures but various functions[35]. Epigenetic mechanism in regulation of gene expression in the SERPIN family was first shown inSERPINB5(maspin)[22]. Both hyper- and hypo-methylation of theSERPINB5gene has been found to be involved in up- and down-regulation in many types of human carcinomas[36-43]. For the heat- shock protein 47 (Hsp47) encoding gene,SERPINH1, it was also found that the methylation inversely correlates with gene expression in human neuroblastoma cells[44]. The acetylation of histones H3 and H4 as well as methylation of Lys4 of histone H3 are correlated with the tissue-specific expression of a cluster of serpin genes at chromosome 14q32.1, includingSERPINA3(α1-antichymotrypsin),SERPINA4(kallistatin) andSERPINA5(protein C inhibitor)[45]. Chelbi et al.[23], reported methylation profiles for 18 human serpin genes in preeclampsia-a pregnancy-induced hypertensive disorder. Most of serpin promoters were demonstrated to be either totally methylated or totally unmethylated, whereasSERPINA3,A5andA8displayed complex methylation profiles and position-specific methylation may correlate with mRNA expression level. In contrast, no such information forSERPINE1(PAI-1) andSERPINE2(PN-1) has been provided. We have previously demonstrated the DNA methylation at 5’ flanking region of PAI-1 gene is inversely correlated with the PAI-1 mRNA level in a number of human cell lines[24]. However, DNA methylation was less important in the regulation of the PAI-1 gene expression in human oral carcinoma patients, especially in the normal oral mucosa from healthy volunteers[20]. For ratSERPINE2,in vitroCpG methylation could block transcription from the PN-1 promoter in rat hepatoma cells but not in C6 rat glioma cells, indicating methylation related regulation with cell-specific pattern. Cortese et al.[45]reported that nine genes show inverse correlation between methylation and gene expression in human fetal lung and adult lung, includingSERPINE2, but that there was no such correlation when comparing normal and cancer tissues. The region of theSERPINE2gene analysed by those authors includes only a fragment upstream of the CpG island which we analysed (personal communication )[46]. Therefore, it is impossible to compare their results with ours.
Conclusively, we have demonstrated that CpG methylation and histone deacetylation may regulate PN-1 mRNA expression, and methylation at specific sites to some extent inversely correlates with PN-1 expression but the effect of methylation inhibitors is most likely indirect. Other unknown factors need to be further investigated for their determining roles in the highly variable expression among different cell types.
REFERENCES
1. Andreasen PA, Kjoller L, Christensen L, et al. The urokinase-type plasminogen activator system in cancer metastasis: a review. Int J Cancer 1997; 72: 1-22.
2. Andreasen PA, Egelund R, Petersen HH. The plasminogen activation system in tumor growth, invasion, and metastasis. Cell Mol Life Sci 2000; 57: 25-40.
3. Durand MK, Bodker JS, Christensen A, et al. Plasminogen activator inhibitor-I and tumor growth, invasion, and metastasis. Thromb Haemost 2004; 91: 438-49.
4. Duffy MJ. The urokinase plasminogen activator system: role in malignancy. Curr Pharm Des 2004; 10: 39-49.
5. Harris L, Fritsche H, Mennel R, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol 2007; 25: 5287-312.
6. Fong KM, Kida Y, Zimmerman PV, et al. TIMP1 and adverse prognosis in non-small cell lung cancer. Clin Cancer Res 1996; 2: 1369-72.
7. Kallakury BV, Karikehalli S, Haholu A, et al. Increased expression of matrix metalloproteinases 2 and 9 and tissue inhibitors of metalloproteinases 1 and 2 correlate with poor prognostic variables in renal cell carcinoma. Clin Cancer Res 2001; 7: 3113-9.
8. Nakopoulou L, Giannopoulou I, Lazaris ACh, et al. The favorable prognostic impact of tissue inhibitor of matrix metalloproteinases-1 protein overexpression in breast cancer cells. APMIS 2003; 111: 1027-36.
9. Scrideli CA, Cortez MA, Yunes JA, et al. mRNA expression of matrix metalloproteinases (MMPs) 2 and 9 and tissue inhibitor of matrix metalloproteinases (TIMPs) 1 and 2 in childhood acute lymphoblastic leukemia: potential role of TIMP1 as an adverse prognostic factor. Leuk Res 2010; 34: 32-7.
10. Duffy MJ, Crown J. A personalized approach to cancer treatment: how biomarkers can help. Clin Chem 2008; 54: 1770-9.
11. Duffy MJ, McGowan PM, Gallagher WM. Cancer invasion and metastasis: changing views. J Pathol 2008; 214: 283-93.
12. Buchholz M, Biebl A, Neesse A, et al. SERPINE2 (protease nexin I) promotes extracellular matrix production and local invasion of pancreatic tumors in vivo. Cancer Res 2003; 63: 4945-51.
13. Selzer-Plon J, Bornholdt J, Friis S, et al. Expression of prostasin and its inhibitors during colorectal cancer carcinogenesis. BMC Cancer 2009; 9: 201.
14. Candia BJ, Hines WC, Heaphy CM, et al. Protease nexin-1 expression is altered in human breast cancer. Cancer Cell Int 2006; 6: 16.
15. Gao S, Krogdahl A, Sorensen JA, et al. Overexpression of protease nexin-1 mRNA and protein in oral squamous cell carcinomas. Oral Oncol 2008; 44: 309-13.
16. Nagahara A, Nakayama M, Oka D, et al. SERPINE2 is a possible candidate promotor for lymph node metastasis in testicular cancer. Biochem Biophys Res Commun 2010; 391: 1641-6.
17. Offersen BV, Nielsen BS, Hoyer-Hansen G, et al. The myofibroblast is the predominant plasminogen activator inhibitor-1-expressing cell type in human breast carcinomas. Am J Pathol 2003; 163: 1887-99.
18. Illemann M, Hansen U, Nielsen HJ et al. Leading-edge myofibroblasts in human colon cancer express plasminogen activator inhibitor-1. Am J Clin Pathol 2004; 122: 256-65.
19. Usher PA, Thomsen OF, Iversen P et al. Expression of urokinase plasminogen activator, its receptor and type-1 inhibitor in malignant and benign prostate tissue. Int J Cancer 2005; 113: 870-80.
20. Gao S, Nielsen BS, Krogdahl A et al. Epigenetic alterations of the SERPINE1 gene in oral squamous cell carcinomas and normal oral mucosa. Genes Chromosomes Cancer 2010; 49: 526-38.
21. Lindberg P, Larsson A, Nielsen BS. Expression of plasminogen activator inhibitor-1, urokinase receptor and laminin gamma-2 chain is an early coordinated event in incipient oral squamous cell carcinoma. Int JCancer 2006; 118: 2948-56.
22. Futscher BW, Oshiro MM, Wozniak RJ et al. Role for DNA methylation in the control of cell type specific maspin expression. Nat Genet 2002; 31: 175-9.
23. Chelbi ST, Mondon F, Jammes H et al. Expressional and epigenetic alterations of placental serine protease inhibitors: SERPINA3 is a potential marker of preeclampsia. Hypertension 2007; 49: 76-83.
24. Gao S, Skeldal S, Krogdahl A, et al. CpG methylation of the PAI-1 gene 5'-flanking region is inversely correlated with PAI-1 mRNA levels in human cell lines. Thromb Haemost 2005; 94: 651-60.
25. Erno H, Monard D. Molecular organization of the rat glia-derived nexin/protease nexin-1 promoter. Gene Expr 1993; 3: 163-74.
26. Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol 1987; 196: 261-82.
27. Guttridge DC, Cunningham DD. Characterization of the human protease nexin-1 promoter and its regulation by Sp1 through a G/C-rich activation domain. J Neurochem 1996; 67: 498-507.
28. Shen L, Guo Y, Chen X, et al. Optimizing annealing temperature overcomes bias in bisulfite PCR methylation analysis. Biotechniques 2007; 42: 48, 50, 52.
29. Wong WK, Chen K, Shih JC. Decreased methylation and transcription repressor Sp3 up-regulated human monoamine oxidase (MAO) B expression during Caco-2 differentiation. J Biol Chem 2003; 278: 36227-35.
30. Zhu WG, Srinivasan K, Dai Z, et al. Methylation of adjacent CpG sites affects Sp1/Sp3 binding and activity in the p21(Cip1) promoter. Mol Cell Biol 2003; 23: 4056-65.
31. Zhao H, Shiina H, Greene KL, et al. CpG methylation at promoter site -140 inactivates TGFbeta2 receptor gene in prostate cancer. Cancer 2005; 104: 44-52.
32. Furuta T, Shuto T, Shimasaki S, et al. DNA demethylation-dependent enhancement of toll-like receptor-2 gene expression in cystic fibrosis epithelial cells involves SP1-activated transcription. BMC Mol Biol 2008; 9: 39.
33. Miyajima A, Furihata T, Chiba K. Functional analysis of GC Box and its CpG methylation in the regulation of CYP1A2 gene expression. Drug Metab Pharmacokinet 2009; 24: 269-76.
34. Zong J, Ashraf J, Thompson EB. The promoter and first, untranslated exon of the human glucocorticoid receptor gene are GC rich but lack consensus glucocorticoid receptor element sites. Mol Cell Biol 1990; 10: 5580-5.
35. Law RH, Zhang Q, McGowan S, et al. An overview of the serpin superfamily. Genome Biol 2006; 7: 216.
36. Akiyama Y, Maesawa C, Ogasawara S, et al. Cell-type-specific repression of the maspin gene is disrupted frequently by demethylation at the promoter region in gastric intestinal metaplasia and cancer cells. Am J Pathol 2003; 163: 1911-9.
37. Domann FE, Rice JC, Hendrix MJ, et al. Epigenetic silencing of maspin gene expression in human breast cancers. Int J Cancer 2000; 85: 805-10.
38. Fujisawa K, Maesawa C, Sato R, et al. Epigenetic status and aberrant expression of the maspin gene in human hepato-biliary tract carcinomas. Lab Invest 2005; 85: 214-24.
39. Murakami J, Asaumi J, Maki Y, et al. Effects of demethylating agent 5-aza-2(')-deoxycytidine and histone deacetylase inhibitor FR901228 on maspin gene expression in oral cancer cell lines. Oral Oncol 2004; 40: 597-603.
40. Ogasawara S, Maesawa C, Yamamoto M, et al. Disruption of cell-type-specific methylation at the Maspin gene promoter is frequently involved in undifferentiated thyroid cancers. Oncogene 2004; 23: 1117-24.
41. Rose SL, Fitzgerald MP, White NO, et al. Epigenetic regulation of maspin expression in human ovarian carcinoma cells. Gynecol Oncol 2006; 102: 319-24.
42. Sato N, Fukushima N, Matsubayashi H, et al. Identification of maspin and S100P as novel hypomethylation targets in pancreatic cancer using global gene expression profiling. Oncogene 2004; 23: 1531-8.
43. Yatabe Y, Mitsudomi T, Takahashi T. Maspin expression in normal lung and non-small-cell lung cancers: cellular property-associated expression under the control of promoter DNA methylation. Oncogene 2004; 23: 4041-9.
44. Yang Q, Liu S, Tian Y, et al. Methylation-associated silencing of the heat shock protein 47 gene in human neuroblastoma. Cancer Res 2004; 64: 4531-8.
45. Gopalan SM, Wilczynska KM, Konik BS, et al. Astrocyte-specific expression of the alpha1-antichymotrypsin and glial fibrillary acidic protein genes requires activator protein-1. J Biol Chem 2006; 281: 1956-63.
46. Cortese R, Hartmann O, Berlin K, et al. Correlative gene expression and DNA methylation profiling in lung development nominate new biomarkers in lung cancer. Int J Biochem Cell Biol 2008; 40: 1494-508.
10.1007/s11670-011-0092-5
2010-11-25;Accepted2011-03-11
This work was supported by the Danish National Research Foundation (26-331-6); the Danish Cancer Society (DP 07043, DP 08001); Grosserer Alfred Nielsen and Hustrus Fond.
*Corresponding author.
E-mail: shg@mb.au.dk
© Chinese Anti-cancer Association and Springer-Veriag Berlin Heidelberg 2011
杂志排行
Chinese Journal of Cancer Research的其它文章
- Procyanidins Inhibit Tumor Angiogenesis by Crosslinking Extracellular Matrix
- Cytochrome P450 2E1 RsaI/PstI and DraI Polymorphisms Are Risk Factors for Lung Cancer in Mongolian and Han Population in Inner Mongolia
- Phase I Study to Determine MTD of Docetaxel and Cisplatin with Concurrent Radiation Therapy for Stage III Non-SmallCell Lung Cancer
- Breast Cancer Subtypes and Survival in Chinese Women with Operable Primary Breast Cancer
- γ-Secretase Inhibitor, DAPT Inhibits Self-renewal and Stemness Maintenance of Ovarian Cancer Stem-like Cells In Vitro
- Over-expression of Metastasis-associated in Colon Cancer-1 (MACC1) Associates with Better Prognosis of Gastric Cancer Patients
