Wild-Type KRAS and BRAF Could Predict Response to Cetuximab in Chinese Colorectal Cancer Patients
2011-07-18JingGaoTingtingWangJingweiYuYanyanLiLinShen
Jing Gao, Ting-ting Wang, Jing-wei Yu, Yan-yan Li, Lin Shen
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Department of Gastrointestinal Oncology, Peking University School of Oncology, Beijing Cancer Hospital & Institute, Beijing 100142, China
Wild-Type KRAS and BRAF Could Predict Response to Cetuximab in Chinese Colorectal Cancer Patients
Jing Gao, Ting-ting Wang, Jing-wei Yu, Yan-yan Li, Lin Shen*
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Department of Gastrointestinal Oncology, Peking University School of Oncology, Beijing Cancer Hospital & Institute, Beijing 100142, China
Objective: To analyze the relationship between KRAS, BRAF mutations and the response to Cetuximab in Chinese colorectal cancer patients.
Methods: A total of 273 Chinese colorectal cancer patients were evaluated for KRAS and BRAF mutations by Sanger sequencing. Among them, 59 patients with metastatic colorectal cancer (mCRC) were treated with Cetuximab in combination with chemotherapy from August 2005 to July 2009. Statistical analysis was conducted to assess the relationship between KRAS, BRAF mutations and the response or survival of 59 mCRC patients.
Results: KRAS and BRAF mutation rates were 38.5% (105/273) and 5.1% (14/273), respectively, and KRAS/BRAF mutations were mutually exclusive. Among 59 patients treated with Cetuximab plus chemotherapy, KRAS and BRAF mutations were identified in 11 and 5 patients, respectively. The response rates and median progression-free survivals (PFS) in KRAS wild-type and mutant patients were 35.4% (17/48) vs. 9.1% (1/11) (P=0.054) and 153 days vs. 99 days (P=0.01), respectively. Also, the response rates and median PFS in BRAF wild-type and mutant patients were 37.2% (16/43) vs. 20% (1/5) (P=0.016) and 138 days vs. 90 days (P=0.036), respectively.
Conclusion: Besides KRAS, assessing BRAF mutation should also be required to select patients eligible for Cetuximab. Further prospective evaluation in large samples should be performed to confirm these preliminary findings.
KRAS; BRAF; Mutation; Cetuximab; Colorectal cancer
INTRODUCTION
The mortality and morbidity of colorectal cancer (CRC) in China are increasing day by day, and CRC becomes the third-leading cause of cancer-related deaths in China. Although the standard combination chemotherapy regimens containing 5-fluorouracil, oxaliplatin or irinotecan had brought significant improvement for CRC patients, the 5-year survival rate of CRC was still low[1]. Recently, Cetuximab, one monoclonal antibody targeting epidermal growth factor receptor (EGFR), has been proved to be effective in about 10%-20% CRC patients[2,3]. Cetuximab plays its role through binding to EGFR and inhibiting the downstream RAS/RAF/MEK/ERK or PI3K/PTEN/AKT signal pathways, but no association was seen between EGFR expression and Cetuximab efficacy[4,5].
KRAS and BRAF are two of molecules downstream iiiiiiiiiiiof EGFR and play an important role in EGFR signaling cascade. The activating mutations in exon 2 of KRAS can induce unlimited proliferation of tumor cells, thus isolating the pathway from the control of EGFR[6,7]. Others have reported that KRAS mutations could become an independent prognostic factor in advanced CRC patients treated with Cetuximab, and patients with mutated KRAS could not benefit from Cetuximab[8,9]. So, detection of KRAS mutations is strongly recommended by FDA before administration of Cetuximab. Nevertheless, not all patients with wild-type KRAS had a response to Cetuximab, maybe other molecules downstream of EGFR could affect the response to Cetuximab.
Phosphatase and tensin (PTEN) is one of downstream molecules of EGFR, and the loss of PTEN protein can stimulate the proliferation of cancer cells[10]. Studies in western populations have demonstrated that positive PTEN expression could predict the response to Cetuximab[11], which was consistent with the result performed in Chinese CRC patients[12]. As another downstream molecule of EGFR, BRAF also plays animportant role in CRC. BRAF mutation in western CRC patients was associated with poor prognosis, and patients with BRAF mutation could not respond to Cetuximab or had shorter progression-free survival (PFS) and shorter overall survival (OS)[13-16]. However, according to recent analysis from prospective data in CRYSTAL trial, BRAF status could not predict the benefit of addition of Cetuximab, and only as a predictive marker (paper presented in 2010 ASCO GI, not published). These results indicated that BRAF mutation as a predictive marker is controversial and more important as a prognostic marker. Up to now, little is known about the correlation between BRAF mutation and the activity of Cetuximab in Chinese CRC patients. So, this study was conducted to evaluate simultaneously the associations between KRAS, BRAF mutations and the response to Cetuximab in Chinese metastatic CRC (mCRC) patients.
MATERIALS AND METHODS
Patients and Mutation Analysis
A total of 273 histologically confirmed CRC patients with primary tumor tissues treated in Beijing Cancer Hospital from August 2005 to July 2009 were evaluated for KRAS and BRAF mutations in this study. Genomic DNA of formalin-fixed, paraffin-embedded (FFPE) sections with ≥50% tumor cells (if the content of tumor cells in sections was lower than 50%, the sections would be microdissected) was extracted using E.Z.N.A.FFPE DNA Kit (Lot. D3399-01, OMEGA, USA) according to the manufacturer’s instructions. All genomic DNAs were stored at -20°C until further research. A DNA fragment including codons 12 and 13 in exon 2 of KRAS gene was amplified by PCR using primers (KRAS-F: 5’-GGTACTGGTGGAGTATTTGATAG-3’, KRAS-R: 5’-TGGTCCTGCACCAGTAATATG-3’) with a product size of 248 bp. Another DNA fragment including exon 15 of BRAF gene was amplified by PCR using primers (BRAF-F: 5’-CTCTTCATAATGCTTGCTCTGATAGG-3’, BRAF-R: 5’-GTGGAAAAATAGCCTCAATTCTTACC-3’) with a product size of 211 bp. Each PCR reaction consisted of 10× LA PCR buffer II 2 µl, 10 mmol/L dNTPs 2 µl, LA Taq 0.2 µl (DRR200A, TAKARA), genomic DNA 2 µl, 10 µmol/L forward primer 0.5 µl, and 10 µmol/L reverse primer 0.5 µl in a final volume of 20 µl. The cycling conditions were 94°C for 5 min, 45 cycles of 94°C for 30 s, 56°C for 30 s and 72°C for 20 s, final extension at 72 °C for 10 min, and ended at 4 °C. The PCR products were determined by 3% agarose gel electrophoresis and then sequenced using the same forward primer of each gene by Invitrogen 3730XL genetic analyzer. The sequencing results were analyzed with Chromas software under the condition of signal/noise >98%.
Treatment of Patients
Among 273 patients, 59 patients with mCRC were treated with weekly Cetuximab (400 mg/m2as an initial loading dose, and 250 mg/m2subsequent dose) in combination with chemotherapy (standard dose): 34 patients received Cetuximab plus oxaliplatin-based chemotherapy, 22 patients received Cetuximab plus irinotecan-based chemotherapy, and 3 patients received Cetuximab plus fluorouracil chemotherapy. Cetuximab was administered as first-line treatment in 27 patients, as second-line treatment in 14 patients, and as third-line or more treatment in 18 patients.
Response Evaluation and Survival of Patients
Tumor response was estimated every two months by computed tomography (CT) according to the Response Evaluation Criteria in Solid Tumors (RECIST)[17]. Patients were categorized by complete response (CR), partial response (PR), stable disease (SD), and progressive disease (PD). The overall response rate was defined as (CR+PR)/Total, and the PFS was calculated from the initial treatment of Cetuximab to disease progression or death from any cause. The OS of patients was calculated from the initial treatment of Cetuximab to death from any cause.
Statistical Analysis
The relationships between KRAS, BRAF mutations and the response to Cetuximab were performed using Fisher’s exact test. Kaplan-Meier curves and log-rank test were used to analyze the association between KRAS, BRAF mutations and PFS or OS, with its associated 95% confidence interval (95% CI). Statistical analysis was done using SPSS 13.0 (SPSS Inc, Chicago, Illinois, USA).P<0.05 represents significant difference.
RESULTS
Patient Characteristics
Of all patients, the median age was 59 years (range 22-84 years) with 160 males and 113 females. Among 59 patients treated with Cetuximab, the median age was 57 years (range 24–81 years) with 38 males and 21 females. The primary locations of tumors were colon (n=25) and rectum (n=33), and one patients with primary locations both at colon and at rectum. More details are shown in Table 1.
KRAS and BRAF Mutational Profiling
Of the 273 CRC patients, 105 patients harbored KRAS mutations and 14 patients harbored BRAF mutations. The mutation types of KRAS in this study included G12D (43/105, 41.0%), G12V (20/105, 19.0%), G12S (9/105, 8.6%), G12C (8/105, 7.6%), G12A (7/105, 6.7%), G12R (1/105, 1.0%), and G13D (17/105, 16.2%). All BRAF mutations were V600E. Wild-type and representative mutant KRAS and BRAF profiles are shown in Figure 1. KRAS and BRAF mutations were mutually exclusive. Among 59 patients treated with Cetuximab, 11 patients harbored KRAS mutations (8 patients with codon 12 mutations, 3 patients with codon 13 mutations) and 5 patients harbored BRAF V600E mutation.
KRAS/BRAF Mutations and the Response or Survival
Up to July 2010, the median OS had not been obtained because only 12 patients died. Of all patients, patients with PR, SD and PD were 18, 21 and 20, respectively. Among 48 KRAS wild-type patients treated with Cetuximab, patients with PR, SD and PD were 17, 18 and 13, respectively, while among 11 KRAS mutant patients, patients with PR, SD and PD were 1, 3 and 7, respectively (P=0.054, Table 2). The median PFS in KRAS wild-type and KRAS mutant patients were 153 days (95% CI: 117.55-188.45) and 99 days (95% CI: 31.80-166.20), respectively (P=0.01, Figure 2A).

Table 1. Patient Characteristics
Among 48 KRAS wild-type patients, 5 patients were identified to carry BRAF mutation. Patients with PR, SD and PD among 43 BRAF wild-type patients and 5 BRAF mutant patients were 16, 18, 9 and 1, 0, 4, respectively (P=0.016, Table 2). The median PFSs in BRAF wild-type and BRAF mutant patients were 138 days (95% CI: 116.07-213.93) and 90 days (95% CI: 57.79-122.21), respectively (P=0.036, Figure 2B).
Of all 59 patients, 43 patients carried neither KRAS nor BRAF mutations, and 16 patients carried either mutant KRAS or mutant BRAF. Patients with PR, SD and PD among 43 wild-type patients and 16 mutant patients were 16, 18, 9 and 2, 3, 11, respectively (P=0.003, Table 2). The median PFS in wild-type and mutant patients were 165 days (95% CI: 116.07-213.93) and 90 days (95% CI: 47.83-132.17), respectively (P<0.001, Figure 2C).
Among 59 patients treated with Cetuximab in combination with chemotherapy, the median PFSs of patients with PR (n=18), SD (n=21) and PD (n=20) were 187 days (95% CI: 139.03-234.97), 159 days (95% CI: 71.35-246.65), and 78 days (95% CI: 53.90-102.11) (P<0.001, Figure 3).
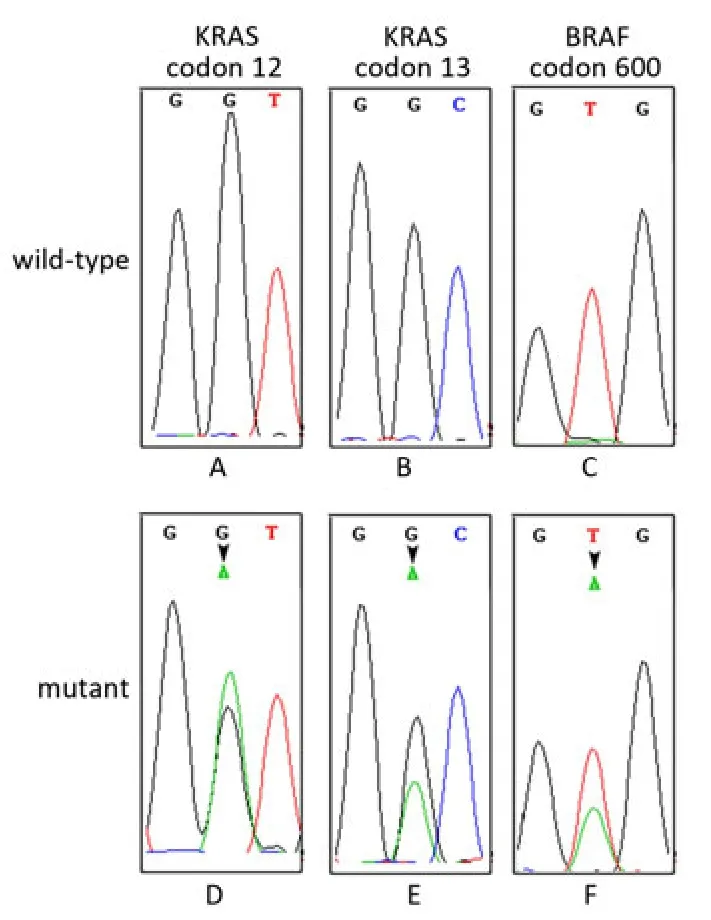
Figure 1. Representative sequencing results showing KRAS and BRAF genotypes. A, B, and C represent wild-type KRAS and BRAF, respectively; D, E, and F represent mutant KRAS and BRAF.
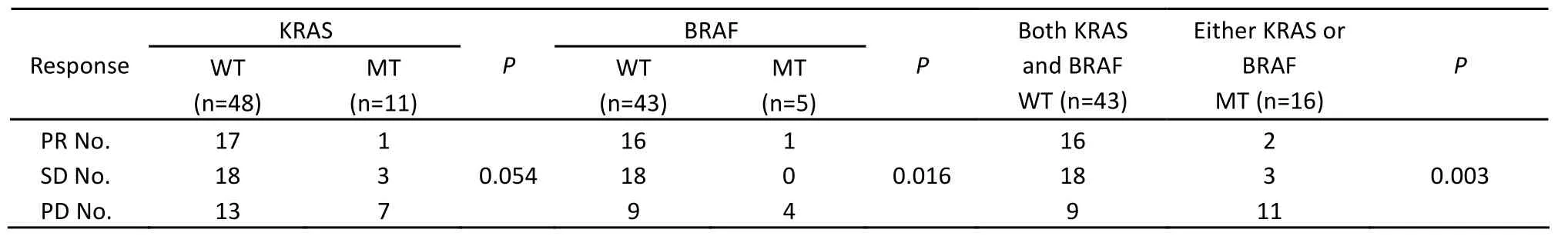
Table 2. Patients’ clinical response in different groups
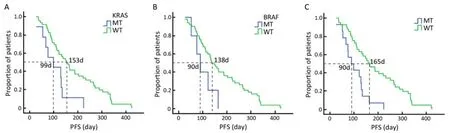
Figure 2. PFS curves of patients with different genotypes. WT: wild-type; MT: mutant.

Figure 3. PFS curves of patients with PR, SD and PD.
DISCUSSION
Of 273 CRC patients, 105 patients harbored KRAS mutations (38.5%) with males 54 (54/160, 33.8%) and females 51 (51/113, 45.1%). Although there was not significant difference between males and females (P=0.06) in this study, KRAS mutation rate in females was higher than that in males. Moreover, the overall KRAS mutation rate in this study was consistent with those in western CRC patients[18]or in another Chinese population[12]. Fourteen patients harbored BRAF mutation (14/273, 5.1%) in our study. Because KRAS and BRAF mutations were mutually exclusive, BRAF mutation rate in KRAS wild-type patients was 8.3% (14/168), which was similar to others[19,20].
Cetuximab is a chimeric mouse/human monoclonal antibody against EGFR, and the development of Cetuximab has brought new expectation to CRC patients. Evidences indicated that the effect of Cetuximab was tightly associated with KRAS mutation status, so the US Food and Drug Administration recommended that patients should undergo KRAS mutation analysis before receiving Cetuximab treatment.
Not all patients with wild-type KRAS could benefit from Cetuximab and no association was seen between EGFR expression and Cetuximab efficacy, therefore, studies were investigated to focus on the molecules downstream of EGFR signal pathways. As the molecules downstream of EGFR, PTEN and BRAF belong to PI3K/PTEN/AKT and RAS/RAF/MEK/ERK signal pathways, respectively. Positive PTEN expression could predict the response to Cetuximab in western and Chinese CRC patients[11,12], and patients with BRAF mutation in western CRC patients could not respond to Cetuximab and the prognosis of them was poor[13-16]. Little is known about the correlation between BRAF mutation and the activity of Cetuximab in Chinese CRC patients.
We analyzed the associations between KRAS, BRAF mutations and the response to Cetuximab in Chinese mCRC patients. Eleven and 5 patients out of 59 mCRC patients carried KRAS mutations and BRAF mutations, respectively. The mutation rate of KRAS in 59 patients was only 18.6% (11/59) which was lower than the overall mutation rate (38.5%), because part of patients received Cetuximab treatment after learning of wild-type KRAS.
From our results, over 20% patients had progressive disease after treated with Cetuximab, which indicated that other factors also influenced the effect of Cetuximab. Besides the mutations of codon 12 and 13 of KRAS, there were other mutations, such as codon 61 and 146. Study reported that KRAS codon 61 and 146 mutations could predict resistance to Cetuximab in KRAS codon 12 and 13 wild-type CRC patients[14]. Besides BRAF V600E mutation of exon 15 (mutation rate over 90% among all BRAF mutations), exon 11 was another mutational hot spot[21]. Although exon 11 mutations of BRAF maybe play some role in the activity of Cetuximab, the mutation rate was very low and it was very difficult to understand the correlation between BRAF exon 11 mutations and the efficacy of Cetuximab. Since Cetuximab plays its role through inhibit EGFR signal pathways, any molecule involved in EGFR signal pathways maybe influence the activity of Cetuximab.
Figure 3 in this study showed the median PFS in patients with PR was slightly longer than that in patients with SD, but there was not significant difference between them. Previous study classified patients with SD to nonresponders of Cetuximab[15], maybe in the futurestudy, patients with SD should also be classified to responders.
In conclusion, we analyzed the associations between KRAS, BRAF mutations and the response to Cetuximab in Chinese mCRC patients. The results demonstrated that both KRAS and BRAF mutations were inversely associated with the response and the PFS to Cetuximab. Combined KRAS and BRAF mutations should be assessed to select patients eligible for Cetuximab. In the future study, we’ll confirm these results in a large number of patients.
Acknowledgement
The authors thank Dr. Ji-ping Yue (Infections and Cancer Biology Group, International Agency for Research on Cancer, Lyon, France) for critical reading of the manuscript.
REFERENCES
1. Venook AP. Epidermal growth factor receptor targeted treatment for advanced colorectal carcinoma. Cancer 2005; 103:2435-46.
2. Saltz LB, Meropol NJ, Loehrer PJ Sr, et al. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol 2004; 22:1201-8.
3. Cunningham D, Humblet Y, Siena S, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med 2004; 351:337-45.
4. Chung KY, Shia J, Kemeny NE, et al. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol 2005; 23:1803-10.
5. Hebbar M, Wacrenier A, Desauw C, et al. Lack of useful- ness of epidermal growth factor receptor expression determination for cetuximab therapy in patients with colorectal cancer. Anticancer Drugs 2006; 17:855-7.
6. Forrester K, Almoguera C, Han K, et al. Detection of high incidence of K-ras oncogenes during human colon tumorigenesis. Nature 1987; 327:298-303.
7. Pretlow TP, Brasitus TA, Fulton NC, et al. K-ras mutations in putative preneoplastic lesions in human colon. J. Natl. Cancer Inst 1993; 85:200-4.
8. Lievre A, Bachet JB, Boige V, et al. KRAS mutation as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol 2008; 26:374-9.
9. Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutation and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 2008; 359:1757-65.
10. Sartore-Bianchi A, Martini M, Molinari F, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res 2009; 69: 1851-7.
11. Loupakis F, Pollina L, Stasi I, et al. PTEN expression and KRAS mutations on primary tumors and metastases in the prediction of benefit from cetuximab plus irinotecan for patients with metastatic colorectal cancer. J Clin Oncol 2009; 27: 2622-9.
12. Li FH, Shen L, Li ZH, et al. Impact of KRAS mutation and PTEN expression on cetuximab-treated colorectal cancer. World J Gastroenterol 2010; 16: 5881-8.
13. Richman SD, Seymour MT, Chambers P, et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol 2009; 27:5931-7.
14. Loupakis F, Ruzzo A, Cremolini C, et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer 2009; 101: 715-21.
15. Di Nicolantonio FD, Martini M, Molinari F, et al. Wild-Type BRAF Is Required for Response to Panitumumab or Cetuximab in Metastatic Colorectal Cancer. J Clin Oncol 2008; 26: 5705-12.
16. Laurent-Puig P, Cayre A, Manceau G, et al. Analysis of PTEN, BRAF, and EGFR Status in Determining Benefit From Cetuximab Therapy in Wild-Type KRAS Metastatic Colon Cancer. J Clin Oncol 2009; 27: 5924-30.
17. Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors: European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 2000; 92:205-16.
18. Heinemann V, Stintzing S, Kirchner T, et al. Clinical relevance of EGFR- and KRAS-status in colorectal cancer patients treated with monoclonal antibodies directed against the EGFR. Cancer Treat Rev 2009; 35: 262-71.
19. Rajagopalan H, Bardelli A, Lengauer C, et al. Tumori- genesis: RAF/RAS oncogenes and mismatch-repair status. Nature 2002; 418:934.
20. Yuen ST, Davies H, Chan TL, et al. Similarity of the phenotypic patterns associated with BRAF and KRAS mutations in colorectal neoplasia. Cancer Res 2002; 62:6451-5.
21. Fukushima T, Suzuki S, Mashiko M, et al. BRAF mutations in papillary carcinomas of the thyroid. Oncogene 2003; 22:6455-7.
10.1007/s11670-011-0271-4
2011-03-21; Accepted: 2011-06-17
*Corresponding author.
E-mail: lin100@medmail.com.cn
©Chinese Anti-Cancer Association and Springer-Verlag Berlin Heidelberg 2011
杂志排行
Chinese Journal of Cancer Research的其它文章
- Prognostic Value of Promoter Hypermethylation of Retinoic Acid Receptor Beta (RARB) and CDKN2 (p16/MTS1) in Prostate Cancer
- Expression and Distribution Characteristics of Human Ortholog of Mammalian Enabled (hMena) in Glioma
- Changes of Serum Trace Elements, AFP, CEA, SF, T3, T4 and IGF-Ⅱ in Different Periods of Rat Liver Cancer
- Mast Cells in Adjacent Normal Colon Mucosa rather than Those in Invasive Margin are Related to Progression of Colon Cancer
- Dosimetry Comparison between Volumetric Modulated Arc Therapy with RapidArc and Fixed Field Dynamic IMRT for Local-Regionally Advanced Nasopharyngeal Carcinoma
- Hepatocellular Tumors: Immunohistochemical Analyses for Classification and Prognostication
