A New Problem with Cross-Species Amplification of Microsatellites: Generation of Non-Homologous Products
2010-12-25YUEGenHuaBalazsKovacsLaszloOrban
YUE Gen-Hua, Balazs Kovacs, Laszlo Orban
(1. Reproductive Genomics, Strategic Research Program, Temasek Life Sciences Laboratory, Singapore; 2. Regional University Center of Excellence in Environmental Industry Based on Natural Resources, Gödöllő, Hungary; 3. Department of Biological Sciences, National University of Singapore, Singapore; 4. Molecular Population Genetics Group, Strategic Research Program, Temasek Life Sciences Laboratory, Singapore)
A New Problem with Cross-Species Amplification of Microsatellites: Generation of Non-Homologous Products
YUE Gen-Hua1,4,*, Balazs Kovacs2, Laszlo Orban1,3,*
(1.Reproductive Genomics, Strategic Research Program, Temasek Life Sciences Laboratory, Singapore; 2.Regional University Center of Excellence in Environmental Industry Based on Natural Resources, Gödöllő, Hungary; 3.Department of Biological Sciences, National University of Singapore, Singapore; 4.Molecular Population Genetics Group, Strategic Research Program, Temasek Life Sciences Laboratory, Singapore)
Microsatellites have been widely used in studies on population genetics, ecology and evolutionary biology. However, microsatellites are not always available for the species to be studied and their isolation could be time-consuming. In order to save time and effort researchers often rely on cross-species amplification. We revealed a new problem of microsatellite cross-species amplification in addition to size homoplasy by analyzing the sequences of electromorphs from seven catfish species belonging to three different families (Clariidae, Heteropneustidae and Pimelodidae). A total of 50 different electromorphs were amplified from the seven catfish species by using primers for 4 microsatellite loci isolated from the speciesClarias batrachus. Two hundred and forty PCR-products representing all 50 electromorphs were sequenced and analyzed. Primers for two loci amplified specific products from orthologous loci in all species tested, whereas primers for the other two loci produced specific and polymorphic bands from some non-orthologous loci, even in closely related non-source species. Size homoplasy within the source species was not obvious, whereas extensive size homoplasy across species were detected at three loci, but not at the fourth one. These data suggest that amplification of products from non-orthologous loci and appearance of size homoplasy by cross-amplification are locus dependent, and do not reflect phylogenetic relationship. Amplification of non-orthologous loci and appearance of size homoplasy will lead to obvious complications in phylogenetic interference, population genetic and evolutionary studies. Therefore, we propose that sequence analysis of cross-amplification products should be conducted prior to application of cross-species amplification of microsatellites.
Microsatellite; Polymorphism; Evolution; Non-orthologous loci
Microsatellites are short tandem repeat DNA sequences with the unit length of 1 to 6 base pairs (Weber & May, 1989). Because they are highly polymorphic, co-dominant in nature, easy to score by PCR and rather abundant in most organisms studied, they have been widely used for the study of linkage mapping, comparative mapping, demographic structure and phylogenetic history in populations (Goldstein & Schlotterer, 1999; Zhang et al, 2001). However, microsatellites are not always available for the species to be studied and their isolation could be time-consuming (Lin et al, 2008; Wang et al, 2008). In order to save time and effort researchers often rely on cross-species amplification (Chang et al, 2008; Küpper et al, 2008; Kayser et al, 1996; Kijas et al, 1995; Lin et al, 2008). This procedure uses PCR primers complementary to the flanking regions of loci from a extensively studied (source) species to amplify microsatellites from closely (Harr et al, 1998) or sometimes quite distantly related species (Gonzalez-Martinez et al, 2004) for which no such markers are described. One problem related to cross-species amplification is size homoplasy (Anmarkrud et al, 2008; Estoup et al, 1995). PCR products of microsatellite loci with the same fragment length, but different sequence can arise from mutational events (deletion or insertion) in the flanking regions of the repeats or by interruptions in a perfect repeat producing alleles of the same size, which however are not identical by decent. Microsatellite size homoplasy has been reported in a number of papers (Hempel & Peakall, 2003; Makova et al, 2000; van Oppen et al, 2000) and was thought be a major problem of cross-amplification. It seems that size homoplasy increases with time divergence among populations and taxa (Estoup et al, 1995). However, a current study showed that homoplasy at microsatellite electromorphs did not represent a significant problem for many types of population genetics analyses performed by molecular ecologists, as the extensive variability at microsatellite loci often compensated for their homoplasious evolution (Estoup et al, 2002).
In this paper, we describe a new problem of applying microsatellites for several different taxa. Cross-species amplification of microsatellites generated polymorphic products from non-orthologous loci, which were revealed by sequence analysis of 240 clones representing all 50 electromorphs from four loci in seven species (Clarias batrachus, C. fuscus, C. gariepinus, C. macrocephalus, Heterobranchus longfilis, Heteropneustes fossilisandPhractocephalus hemioliopterus).
1 Materials and Methods
1.1 Species and phylogenetic analyses
Seven species of catfish were used in this study, namely:Clarias batrachus(abbreviation:Cba; the source species),C. fuscus(Cfu),C. gariepinus(Cga),C. macrocephalus(Cma),Heterobranchus longfilis(Hlo),Heteropneustes fossilis(Hfo), andPhractocephalus hemioliopterus(Phe). According to the current taxonomical system, five of the species studied were from the Clariidae family, one (Heteropneustes fossilis) from the Heteropneustidae family, which is closely related to Clariidae and the last (P. hemioliopterus) from the more distant Pimelodidae family. In order to determine the exact evolutionary relationship among the seven catfish species, phylogenetic analyses were conducted on the basis of the partial sequences ofcytbgenes from their mitochondrial genome. The sequences of six speciesC. batrachus[AF235932],C. fuscus[AF416885],C.gariepinus[AF126823],C. macrocephalus[AJ548464],Heterobranchus longfilis[AY995125], andHeteropneustes fossilis[AF126828] were downloaded from Genbank, whereas the one ofP. hemioliopteruswas amplified with PCR and sequenced as described (Agnese & Teugels, 2005). The sequence of the cytb gene of the Asian arowana (Scleropages formosus; DQ023143) was used as an outgroup. All seven sequences were aligned using Clustal_X (Thompson et al, 1997), and a NJ tree was reconstructed using the Kimura-2 parameter model of nucleotide using MEGA 3.0 (Kumar et al, 2001). The partial sequence of the cytb gene ofP. hemioliopteruswas deposited in GenBank under the accession number DQ200272.
1.2 Sequencing of electromorphs generated by cross-species amplification
All 50 electromorphs (Tabs. 1-4) generated in an earlier study (Yue et al, 2003) from four microsatellites (Cba01,Cba03,Cba06andCba20) from each of the seven species were used for cloning and sequencing. PCR products (25 µL) were cleaned using a glassmilk-based optimized procedure described earlier (Yue et al, 2007; Yue & Orban, 2001) prior to ligation of the fragments in to the pGEM-T-Easy vector (Promega) and subsequent transformation into XL-10 gold ultracompetent cells (Stratagene). Colonies were subjected to white/blue selection, and the insert of selected white clones was amplified by colony PCR as described (Yue et al, 2000). Un-incorporated PCR primers were removed by treating 5 µL PCR product for each clone with 0.5 unit shrimp alkalic phosphatase (SAP; USB) and 0.2 unit Exonuclease I (ExoI; USB) in 1× SAP buffer at 37℃ for 30 min, followed by a treatment at 80℃ for 15 min to inactivate the enzymes. One µL treated PCR product was directly used as template for sequencing from both directions using a BigDye kit (Applied Biosystems) and either M13 forward or M13 reverse primer in a PTC-100 PCR machine (MJ Research). Electrophoretic separation of the sequencing products was performed by using an ABI3730xl sequencer (Applied Biosystems). In order to exclude the possibility of cloning artifacts, for each electromorph from each species, multiple clones (at least3) were sequenced. Altogether the following number of clones were sequenced for the four microsatellite types:Cba01–107 clones,Cba03–20 clones,Cba06–50 clones andCba20–63 clones. Alignment of sequences was carried out by using Clustal X (Thompson et al, 1997).
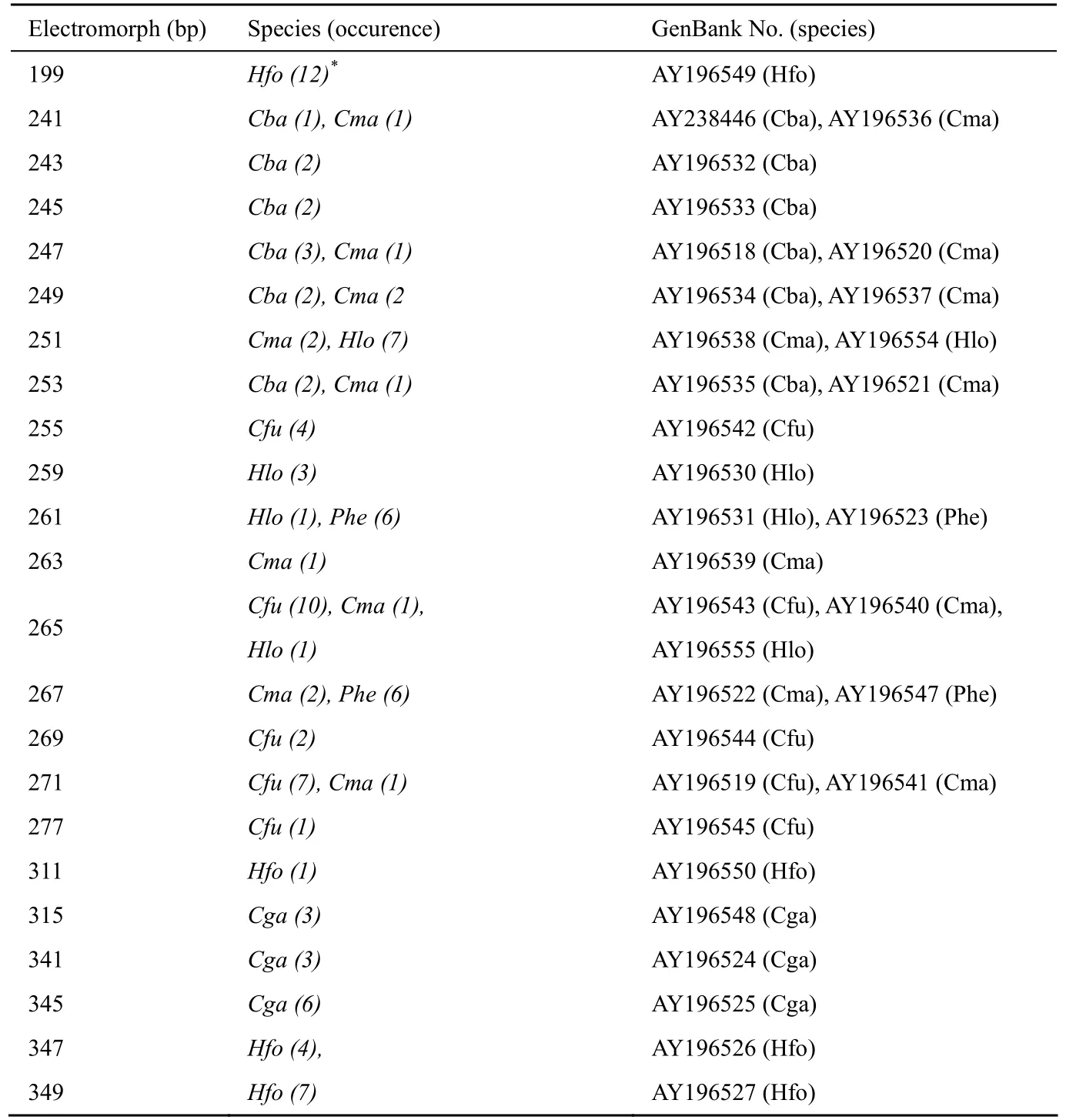
Tab. 1 Electromorphs amplified by the primer pair designed for Cba01 in seven catfish species
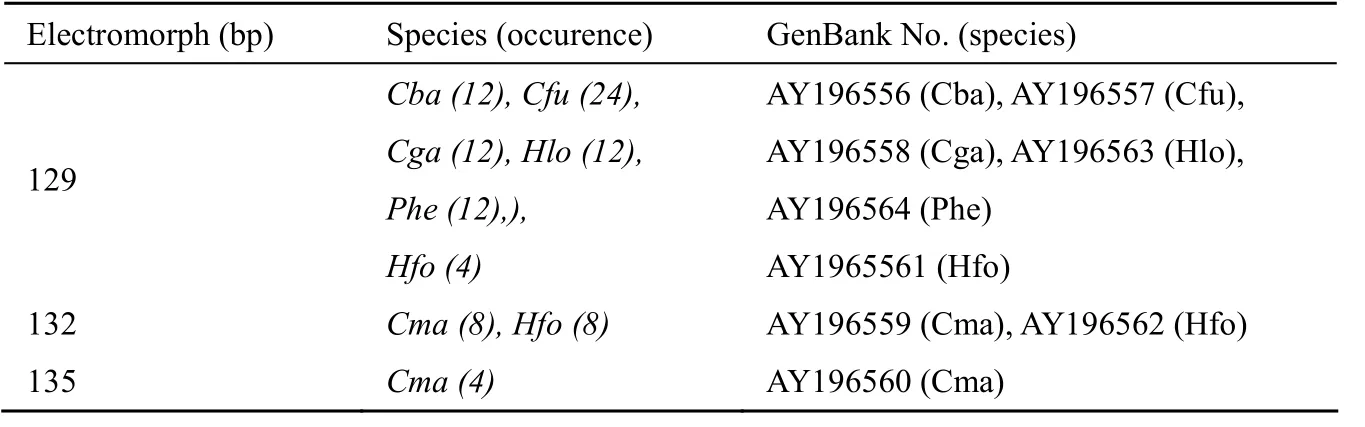
Tab. 2 Electromorphs amplified by the primer pair designed for Cba03 in seven catfish species

Tab. 3 Electromorphs amplified by the primer pair designed for Cba06 in seven catfish species
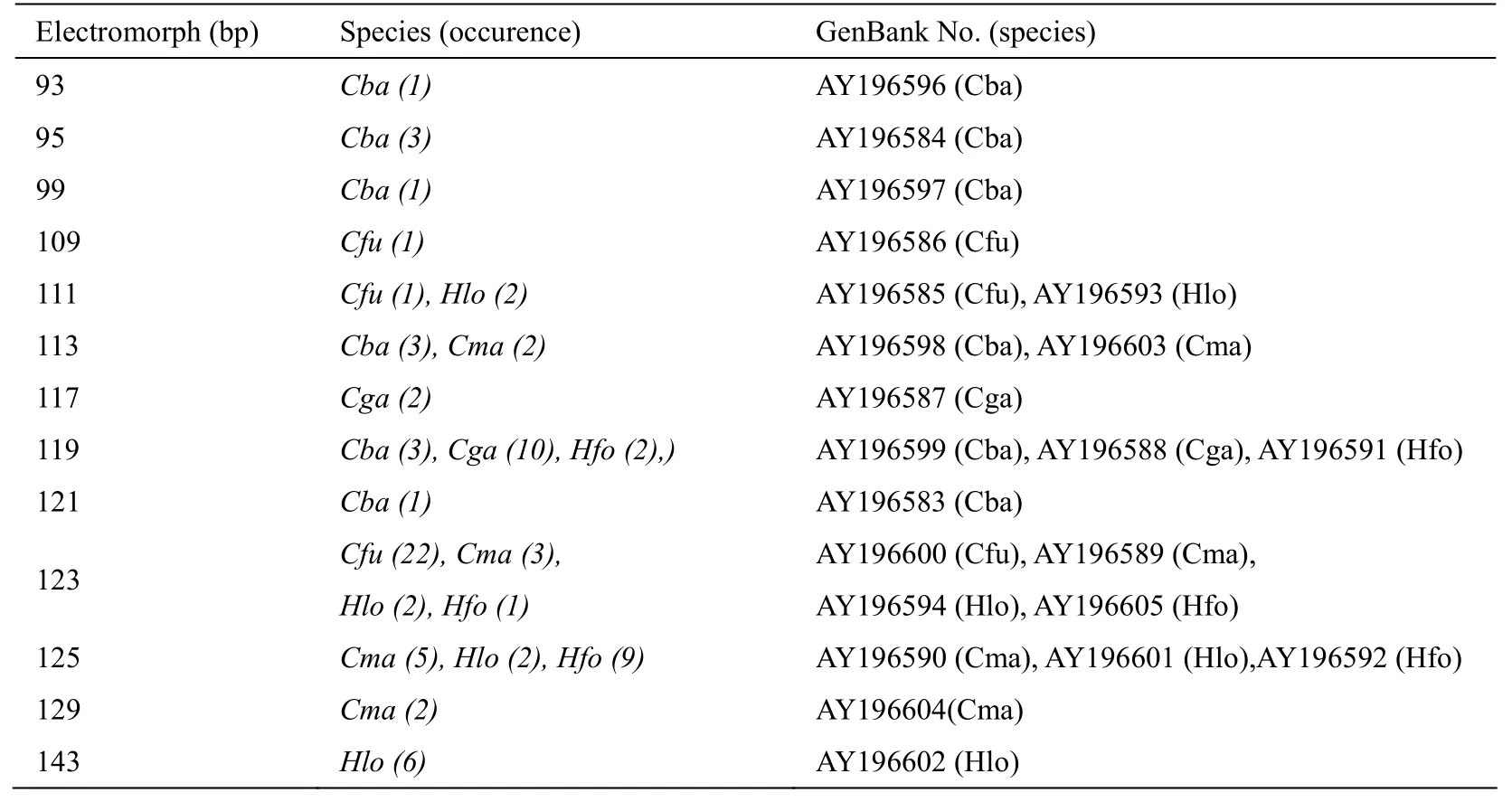
Tab. 4 Electromorphs amplified by the primer pair designed for Cba20 in seven catfish species
2 Results
2.1 Phylogenetic relationship of the seven catfish species
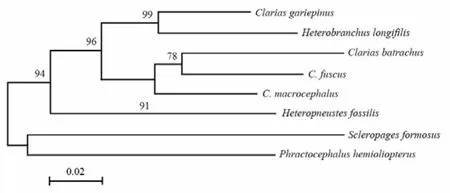
Fig. 1 Phylogenetic relationship among the seven catfish species
Based on the partial sequences of thecytbgene of the seven species, a NJ tree was constructed (Fig. 1). The three speciesClarias batrachus,C. fuscusandC. macrocephaluswere closely related and clustered into a group. This group was linked to the group ofC. gariepinusandHeterobranchus longifilis. The remaining two species:HeteropneustesfossilisandPhractocephalus hemioliopteruswere distantly related to other five species.
2.2 Sequence analysis of electromorphs amplified by the Cba01 primer pair
The primer pair designed to theCba01locus amplified polymorphic products in all seven catfish species tested. Altogether 23 clear bands (eletromorphs) were detected in the seven species (size range: 199-349 bp), their sequencing analyses uncovered the total of 34 different alleles (Tab. 1). InC. fuscus,C. macrocephalusandP. hemioliopterusboth the repeat and the flanking regions exhibited high similarity to source sequences fromC. batrachus(Fig. 2A). On the other hand, the corresponding sequences fromC. gariepinus, andHeteropneustes fossilisspecies were completely different from the source sequences (Fig. 2B), but quite similar among these three species. The length ofHeterobranchus longifilisalleles was similar to those of the source species, but the flanking region and repeats were entirely different (Fig. 2C).
The 5' and 3' flanking sequences for each allele were nearly identical in different individuals ofC. batrachus,C. fuscus,C. macrocephalusandP. hemioliopterus, respectively. On the other hand, several differences were found between sequences from different species both at the 5' and 3' flanking regions (seven and eight positions, respectively). Most of them seem to have been caused by substitution, whereas the rest by insertion or deletion of a single base pair. A notable feature is, that the repeat structures of this locus were slightly different in these four species: (GC)2(AC)nin the source species, (GC)3GT(GC)5-6(AC)5(GC)0-1(AC)ninC. fuscusandP. hemioliopterus, whereas (GC)2-5(AC)0-1(GC)0-4(AC)0-2GC(AC)ninC. macrocephalus(Fig. 2A). Therefore, the polymorphism at this locus was caused by change in the number of either AC or GC repeat units in different species, resulting in fragments of the same length, but with quite different sequences. Within species, size homoplasy could only be detected inC. macrocephalus, but not in the source species,C. fuscusorP. hemioliopterus.

Fig. 2 Sequence alignment of some electromorphs (amplified by the primer pair designed for Cba01) from seven different catfish species
InC. gariepinusandH. fossilis,the sequences of the 7 electromorphs (Tab. 1) were different from those in source species. The flanking sequences were quite similar among different alleles, although the polyA and polyT repeats (located at the 5' and 3' flanking regions, respectively) showed polymorphism both within and among species (Fig. 2B). Moreover, a deletion of 16 bp was detected in the 5' flanking region ofHeteropneustes fossilis(data not shown). InC. gariepinus(but not inH. fossilis) a CAG unit was deleted from the 3' flanking region. A few point mutations, short deletions or insertions have also been detected in the 5' and 3' flanking regions among electromorphs from different species (data not shown). Polymorphism in the repeat at this locus was caused either by a change in the length of polyA stretch in the 5' flanking region, or by the unit number of (GA)n, (GAA)n, (GGA)ncompound repeats or by a deletion of three base pairs CAG and a change in the length of the polyT in the 3' flanking region (Fig. 2B).
InH. fossilis,the locus appeared to be duplicated, because more than two bands were detected in the PCR product of each individual tested, whereas no such phenomenon was observed in the other two species. The 199 bp allele from all six individuals ofH. fossilistested (Genebank No. AY196549) lacked a 150 bp fragment including the 5' flanking region and even the whole repeat region as compared with the largest allele (Hfo349) (Fig. 2B).
InHeterobranchus longifilis, the sequences of electromorphs were entirely differently from the alleles of the source species, although the length of the electromorphs was similar to those of the source species (Fig. 2C). The length polymorphism of the electromorphs was caused by the change of number of CT repeats.
2.3 Sequence analysis of electromorphs amplified by the Cba03 primer pair
At theCba03locus, a total of three electromorphs (range: 129 - 135 bp) were detected across the seven species (Tab. 2). Sequencing of each electromorph (20 clones) revealed that the sequence of this locus was highly conserved across the catfish species studied (Fig. 3). The polymorphism was caused exclusively by the change in the unit number of the (GGA)nrepeat. At three positions of 3' flanking region, single base pair substitution was also seen in two species (C. macrocephalusandP. hemiolopterus). No size homoplasy was identified among individuals of any species.
2.4 Sequence analysis of electromorphs amplified by the Cba06 primer pair
At theCba06locus, a total of 11 electromorphs (range 168 -258 bp) were identified across the seven species (Tab. 3). Their sequence analysis demonstrated that they could be divided into two groups and two individual sequences (Fig.4A - D). Fragments amplified fromC. fuscus(1 allele) andC. macrocephalus(4 alleles) showed an overall high similarity to the source sequence (Fig. 4A). In these two species, an insertion of a 34 bp fragment was detected at the 5' flanking region between the primer and repeats in every allele in comparison to the source sequence. Additional single base pair substitutions, located in the flanking regions were also found. The length polymorphism was caused by the change in the unit number of the (AAC)nrepeat within each species, but among species the length polymorphism could also be caused by change in the extent of polyA in the 3' flanking region or the insertion of a 34 bp fragment into the 5' flanking region. Although no size homoplasy was identified within these two species, its presence was quite obvious among species. For example, the 245 bp electromorph inC. fuscusand that inC. macrocephalusshowed different unit number of CAA-repeats and appearance of a CTA sequence due to an A→T mutation in the latter.

Fig. 3 Sequence alignment of some electromorphs (amplified by the primer pair designed for Cba03) from seven different catfish species shows no size homoplasy within or among species
The second group (Fig. 4B) included sequences fromC. gariepinus(2 alleles), andH. longifilis(1). The DNA sequence of the fragments from the two species showed high similarity to each other, but differed from the source sequence both in their flanking regions and repeat motif [(AAC)nvs. (CA)n]. An insertion of five base pairs (CGAAC) was seen in the 5' flanking region of the speciesH. longifilis, as compared the sequences from theC. gariepinus(Fig. 4B). Apart from this insertion, the length polymorphism was caused by the different number of the (AC)nrepeat units in all fragments. Between the two species, single base pair substitution was observed at several positions of the flanking regions. The 168 bp fragment appeared in both species. However comparison of sequences between the two species revealed two different alleles.
The remaining two sequences (Fig. 4C-D; GenBank Nos. AY196578 and AY196579) originated fromP. hemioliopterusandHeteropneustes fossilis, respectively. They did not show any similarity to the first two groups except the primer binding sites and did not contain repeats.
2.5 Sequence analysis of electromorphs amplified by the Cba20 primer pair

Fig. 4 Sequence alignment of some electromorphs (amplified by the primer pair designed for Cba06) from seven different catfish species
A total of 13 electromorphs (range: 93 - 143 bp) were detected across six species (Tab. 4), but not inP.hemioliopterus. Sequence analysis revealed 10 additional alleles (Fig. 5), without any evidence of homoplasy within the source species. In the 5' flanking region, single base pair substitutions were detected at three positions among species. As compared with the source (4 alleles), sequences fromC. macrocephalus(4) andHeterobranchus longifilis(3) showed an insertion of three basepairs (GTC) in the 3' flanking region. Single basepair substitutions were also detected at two positions of the 3' flanking regions. The repeat region was highly variable within and among species. In the source species, repeat structure for the 95 bp allele was (TC)6GC(TC)2, although longer and shorter alleles showed change in repeat number of longer repeat, the GC(TC)2motif remained constant among all alleles. InC. fuscus(3 alleles), where the (TC)nrepeat was interrupted by a TA unit, the (TC)3upstream from the TA remained unchanged, whereas the downstream (TC)nrepeat showed polymorphism among individuals. InC. gariepinus(2 alleles) the TC repeats were interrupted by GC and TG units at several positions and the polymorphism was caused by the change of the long, upstream TC repeat, whereas the shorter ones remained constant. InC. macrocephalusandHeteropneustesfossilis(3 alleles), the (TC)nrepeat was interrupted by CC, TT and GT motifs, whereas inHeterobranchus longifilisby GC, AG and TG units. The reason for the polymorphism was similar to that described forC. gariepinus.

Fig. 5 Sequence alignment of some electromorphs (amplified by the primer pair Cba20) from seven different catfish species
3 Discussion
Microsatellites are very useful tools for genetic and evolutionary studies. However, their genotyping is based on prior sequence information from the genome to be analyzed. Despite of recent improvements on the procedure (for review see: Zane et al, 2002) the isolation of microsatellites is still cumbersome. One of the possible solutions for this problem is cross-species amplification, which involves the use of primer pairs designed for the flanking region of conserved microsatellites (of a so-called source species) for genotyping in related species amplification (Housley et al, 2006; Kayser et al, 1996; Kijas et al, 1995). Data for several such experiments have been reported in teleosts during the last decade (e.g. Koskinen & Primmer, 1999; Yue et al, 2004; Yue et al, 2003). However all PCR products generated in the non-source species have only been analyzed at the sequence level in a few cases (Kayang et al, 2002; Viard et al, 1998). We have tested the applicability of four conserved microsatellite markers isolated earlier fromC. batrachus(Yue et al, 2003) on six additional catfish species. We found that PCR primer pairs designed for the flanking regions of the fourC. batrachusmicrosatellite loci amplified products in most of the related species. However, sequencing analyses of 240 clones representing 50 electromorphs from seven catfish species revealed a new problem of cross-species amplification of microsatellites: the generation of non-orthologous loci, beside the appearance of size homoplasy. Primer pairs designed for twoC. batrachusloci (Cba03andCba20) amplified highly similar (orthologous) sequence products in all non-source species. On the other hand, those designed for other two loci (Cba01andCba06) yielded polymorphic products with entirely different sequence from some of the distantly related species (e.g.P. hemioliopterusandHeteropneustes fossilis), and even in closely related species (e.g.C. gariepinus) indicating that these bands originated from non-orthologous loci. The amplification of specific products from non-orthologous source was locus-dependent, and did not reflect the phylogenetic relationship. Thus, in the absence of sequence information it would be very difficult to predict whether certain primer pairs will amplify products from orthologous loci in a given non-source species or not. Similar phenomenon was observed earlier in soybean (Peakall et al, 1998) and rice (Chen et al, 2002), but those findings have not been analyzed in detail. Taken together, our data suggest that generation of polymorphic products from non-orthologous loci by cross-species amplification is not a unique feature of certain taxonomic groups in fish, instead it might occur throughout the animal and plant kingdom. Although the mechanisms underlying this phenomenon are not fully understood, they are thought to be related to genome and gene duplication, as well as speciation. Such events are expected be more frequent in fish, since the ancestor of today’s teleosts seems to have experienced an additional round of genome duplication (Meyer & Schartl, 1999; Postlethwait et al, 2000) and chromosome duplications (Chang et al, 2005) after their ancestor has split from that of the other vertebrates. Duplication of microsatellite loci followed by gene conversion can lead to amplification of non-orthologous loci as proposed (Angers et al, 2002).
Sequencing of all alleles of four microsatellite loci in the source and six non-source species showed that length difference of microsatellites was not restricted to their repeat regions. A longer insertion and several shorter insertions were detected in the flanking region of the loci orthologous toCba06in non-source species. At theCba01,Cba06andCba20loci, a number of alleles from different non-source species showed the same length, but with different sequences. At the same time, atCba03locus electromorphs of the same length represented the same sequences, suggesting that size homoplasy for microsatellite markers produced by cross-species amplification is locus-dependent, it does not reflect the phylogenetic relationship. We also found the tendency of increase in the number of interrupted repeats of orthologous loci in non-source species, as observed by others in different taxonomic groups (e.g. Culver et al, 2001; Di Gaspero et al, 2000; Estoup et al, 1995; Garza et al, 1995; van Oppen et al, 2000). This tendency also seems to be locus-dependent in catfish, since two loci (Cba01andCba20) showed clear interruptions in non-source species, whereas the other two (Cba03andCba06) exhibited no or few interruptions in them.
Applications of microsatellites to population genetics, ecological and evolutionary studies rely heavily on the models used for explaining the mutational process of these markers. However, all models relay on the assumption that differences between alleles at orthologous loci are due entirely to changes in the number of repeats. In this study, we demonstrated that appearance of size homoplasy and amplification of non-orthologous products by cross-species amplification were locus-dependent, and did not reflect phylogenetic relationships. Therefore, application of cross amplification of microsatellites to population genetics and phylogenetic analyses in distantly or even in closely related species, might make the interpretation of length difference of electromorphs difficult and cause wrong estimation of evolutionary relationship.
In conclusion, we revealed a new problem of microsatellite cross-species amplification, namely amplification of non-orthologous loci, besides the well-known problem (size homoplasy). The new problem and appearance of size homoplasy will lead to obvious complications for phylogenetic interferences, population genetics, mapping and evolutionary studies. The sequence analysis of products generated by“cross-species primers” should always be performed, as it could reveal previously unrecognized problems and might allow for extracting more information from these loci, thereby increasing their usefulness.
Agnese, JF, Teugels, GG. 2005. Insight into the phylogeny of African Clariidae (Teleostei, Siluriformes): Implications for their body shape evolution, biogeography, and taxonomy [J].Mol Phylogenet Evol, 36 (3): 546-553.
Angers B, Gharbi K, Estoup A. 2002. Evidence of gene conversion events between paralogous sequences produced by tetraploidization in Salmoninae fish [J].J Mol Evol, 54 (4): 501-510.
Anmarkrud JA, Kleven O, Bachmann L, Lifjeld JT. 2008. Microsatellite evolution: Mutations, sequence variation, and homoplasy in the hypervariable avian microsatellite locus HrU10 [J].BMC Evol Biol, 8: 138.
Chang CH, Hsieh LC, Chen TY, Chen HD, Luo L, Lee HC. 2005. Shannon information in complete genomes [J].J Bioinform Comput Biol, 3 (3): 587-608.
Chang YM, Kuang YY, Liang LQ, Lu CY, He JG, Su XW. 2008. Searching for protein-coding genes using microsatellites in common carp by comparing to zebrafish EST database [J].Zool Res, 29 (4): 373-378.
Chen X, Cho YG, McCouch SR. 2002. Sequence divergence of rice microsatellites inOryzaand other plant species [J].Mol GenetGenomics, 268 (3): 331-343.
Culver M, Menotti-Raymond MA, O'Brien SJ. 2001. Patterns of size homoplasy at 10 microsatellite loci in pumas (Puma concolor) [J].Mol Biol Evol, 18 (6): 1151-1156.
Di Gaspero G, Peterlunger E, Testolin R, Edwards KJ, Cipriani G. 2000. Conservation of microsatellite loci within the genusVitis[J].Theor Appl Genet, 101 (1-2): 301-308.
Estoup A, Jarne P, Cornuet JM. 2002. Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis [J].Mol Ecol, 11 (9): 1591-1604.
Estoup A, Tailliez C, Cornuet JM, Solignac M. 1995. Size homoplasy and mutational processes of interrupted microsatellites in two bee species,Apis melliferaandBombus terrestris(Apidae) [J].Mol Biol Evol, 12 (6): 1074-1084.
Garza JC, Slatkin M, Freimer NB. 1995. Microsatellite allele frequencies in humans and chimpanzees, with implications for constraints on allele size [J].Mol Biol Evol, 12 (4): 594-603.
Goldstein DB, Schlotterer C. 1999. Microsatellites: Evolution and Applications [M]. Oxford: Oxford University Press.
Gonzalez-Martinez SC, Robledo-Arnuncio JJ, Collada C, Diaz A, Williams CG, Alia R, Cervera MT. 2004. Cross-amplification and sequence variation of microsatellite loci in Eurasian hard pines [J].Theor Appl Genet, 109 (1): 103-111.
Harr B, Zangerl B, Brem G, Schlotterer C. 1998. Conservation of locus-specific microsatellite variability across species: A comparison of twoDrosophilasibling species,D. melanogasterandD. simulans[J].Mol Biol Evol, 15 (2): 176-184.
Hempel K, Peakall R. 2003. Cross-species amplification from crop soybeanGlycine maxprovides informative microsatellite markers for the study of inbreeding wild relatives [J].Genome, 46 (3): 382-393.
Housley DJ, Zalewski ZA, Beckett SE, Venta PJ. 2006. Design factors that influence PCR amplification success of cross-species primers among 1147 mammalian primer pairs [J].BMC Genomics, 7: 253. Küpper C, Burke T, Székely T, Dawson DA. 2008. Enhanced cross-species utility of conserved microsatellite markers in shorebirds [J].BMC Genomics, 9: 502.
Kayang BB, Inoue-Murayama M, Hoshi T, Matsuo K, Takahashi H, Minezawa M, Mizutani M, Ito S. 2002. Microsatellite loci in Japanese quail and cross-species amplification in chicken and guinea fowl [J].Genet Sel Evol, 34 (2): 233-253.
Kayser M, Ritter H, Bercovitch F, Mrug M, Roewer L, Nurnberg P. 1996. Identification of highly polymorphic microsatellites in the rhesus macaqueMacaca mulattaby cross-species amplification [J].Mol Ecol, 5 (1): 157-159.
Kijas JM, Fowler JC, Thomas MR. 1995. An evaluation of sequence tagged microsatellite site markers for genetic analysis within Citrus and related species [J].Genome, 38 (2): 349-355.
Koskinen MT, Primmer CR. 1999. Cross-species amplification of salmonid microsatellites which reveal polymorphism in European and Arctic grayling, Salmonidae:Thymallusspp [J].Hereditas, 131 (2): 171-176.
Kumar S, Tamura K, Jakobsen IB, Nei M. 2001. MEGA2: molecular evolutionary genetics analysis software [J].Bioinformatics, 17: 1244-1245.
Lin G, Chang A, Yap W, Yue GH. 2008. Characterization and cross-species amplification of microsatellites from the endangered Hawksbill turtle (Eretmochelys imbricate) [J].Conserv Genet, 9: 1071-1073.
Makova KD, Nekrutenko A, Baker RJ. 2000. Evolution of microsatellite alleles in four species of mice (genusApodemus) [J].J Mol Evol, 51 (2): 166-172.
Meyer A, Schartl M. 1999. Gene and genome duplications in vertebrates: the one-to-four (- to-eight in fish) rule and the evolution of novel gene functions [J].Curr Opin Cell Biol, 11 (6): 699-704.
Peakall R, Gilmore S, Keys W, Morgante M, Rafalski A. 1998. Cross-species amplification of soybean (Glycine max) simple sequence repeats (SSRs) within the genus and other legume genera: Implications for the transferability of SSRs in plants [J].Mol Biol Evol, 15 (10): 1275-1287.
Postlethwait JH, Woods IG, Ngo-Hazelett P, Yan YL, Kelly PD, Chu F, Huang H, Hill-Force A, Talbot WS. 2000. Zebrafish comparative genomics and the origins of vertebrate chromosomes [J].Genome Res, 10 (12): 1890-1902.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools [J].Nucleic Acids Res, 25 (24): 4876-4882.
van Oppen MJH, Rico C, Turner GF, Hewitt GM. 2000. Extensive homoplasy, nonstepwise mutations, and shared ancestral polymorphism at a complex microsatellite locus in LakeMalawi cichlids[J].Mol Biol Evol, 17 (4): 489-498.
Viard F, Franck P, Dubois MP, Estoup A, Jarne P. 1998. Variation of microsatellite size homoplasy across electromorphs, loci, and populations in three invertebrate species [J].J Mol Evol, 47 (1): 42-51.
Wang HZ, Yin QQ, Feng ZG, Li, DY, Sun XW, Li C. 2008. Construction of fractional genomic libraries and screening microsatellites DNA ofEsox reiehertiDybowski [J].Zool Res, 29 (3): 245-252.
Weber JL, May PE. 1989. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain-reaction [J].Am J Hum Genet, 44 (3): 388-396.
Yue GH, Chen F, Orban L. 2000. Rapid isolation and characterization of microsatellites from the genome of Asian arowana (Scleropages formosus, Osteoglossidae, Pisces) [J].Mol Ecol, 9 (7): 1007-1009. Yue GH, David L, Orban L. 2007. Mutation rate and pattern of microsatellites in common carp (Cyprinus carpioL.) [J].Genetica, 129 (3): 329-31.
Yue GH, Ho MY, Orban L, Komen J. 2004. Microsatellites within genes and ESTs of common carp and their applicability in silver crucian carp [J].Aquaculture, 234 (1-4): 85-98.
Yue GH, Kovacs B, Orban L. 2003. Microsatellites fromClarias batrachusand their polymorphism in seven additional catfish species [J].Mol Ecol Notes, 3 (3): 465-468.
Yue GH, Orban L. 2001. Rapid isolation of DNA from fresh and preserved fish scales for polymerase chain reaction [J].Mar Biotechnol, 3 (3): 199-204.
Zane L, Bargelloni L, Patarnello T. 2002. Strategies for microsatellite isolation: a review [J].Mol Ecol, 11 (1): 1-16.
Zhang YW, Zhang YP, Aryder O. 2001. Microsatellites and its application [J].Zool Res, 22 (4): 315-320.
微卫星跨物种交叉PCR扩增的一个新问题:扩增非同源产物
岳根华1,4,*, Balazs Kovacs2, Laszlo Orban1,3,*
(1.Reproductive Genomics, Strategic Research Program, Temasek Life Sciences Laboratory, Singapore; 2.Regional University Center of Excellence in Environmental Industry Based on Natural Resources, Gödöllő, Hungary3.Department of Biological Sciences, National University of Singapore, Singapore; 4.Molecular Population Genetics Group, Strategic Research Program, Temasek Life Sciences Laboratory, Singapore)
微卫星已被广泛应用于群体遗传学、生态学和进化生物学研究。然而,一些物种微卫星尚未克隆。为了节省时间和经费,研究人员往往使用一个物种已发表的微卫星引物扩增其近缘物种的微卫星。该研究对属于 3个不同科(Clariidae、Heteropneustidae 和Pimelodidae)的7个鲶鱼物种的微卫星跨物种PCR扩增产物进行了序列分析,研究发现扩增非同源(non-orthologous)产物是微卫星跨物种PCR扩增的一个新问题。该研究共采用4对胡子鲶微卫星座位引物对7个鲶鱼物种进行了跨物种PCR扩增。对获得的204个PCR产物的序列分析结果表明,两对微卫星座位引物扩增了所有7个物种的同源特异产物。而其他两个座位的引物扩增了特异但非同源的多态产物,对近缘物种的扩增也获得类似结果。另外,除胡子鲶等位基因大小异源同型(size homoplasy)的特征不明显外,其他物种在3个微卫星座位都具有这一非常明显的特征。这些数据表明,微卫星跨物种间交叉扩增能产生非同源产物;等位基因大小异源同型与微卫星座位本身有关,而与物种间的亲缘关系无明显的相关性。微卫星跨物种扩增产生的非同源产物和等位基因大小异源同型将使系统发育、群体遗传学和进化研究明显复杂化。因此,在应用微卫星跨物种交叉扩增数据以前,最好对跨物种交叉扩增产物进行测序验证。
2009-09-15;接受日期:2009-12-31
book=132,ebook=192
微卫星;多态性;进化;非同源座位
Q754;Q343.1;Q984.403
A
0254-5853-(2010)02-0131-10
10.3724/SP.J.1141.2010.02131
date: 2009-09-15; Accepted date: 2009-12-31
This study was supported financially by the internal research funding from Temasek Life Sciences Laboratory
*Corresponding authors (通讯作者),Fax: 65-6872-7007,E-mail: genhua@tll.org.sg; laszlo@tll.org.sg
The authors would like to thank Drs. Graham Mair, Arlo Fast, Bela Urbanyi and Lian Chuan Lim, as well as Ferenc Radics, Judit Raczkevi and Gyula Pasareti for supplying fin clips from various catfish species, and the Strategic Research Program of TLL for financial support. B.K. is grateful for the support of the Temasek Life Sciences Laboratory, the Bolyai Research Fellowship of the Hungarian Academy of Sciences, and the Hungarian Scientific Research Fund (OTKA PD79177)..
